For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PGQVRMSAHRAGRPFFARTVRVLAVPIIVFWGLLAVTTNTFIPQVEHVADELAGPMIPNYAPSQVALLHI GEKFQESDSTNLTMLVFEADRPLDDGDHRYYDDLVRRLQADTEHVQYVMDLWGKPITAAGAQSVDGKASF VLLRLAGNIGQLEANESVDAVRDIIADDAPPQGLKVYVSGAAPLASDTLSTANSSLNNITIFTIILIVVM LLLVYRSAACLLVPLPAVLIEMLVAKGVIATLGHFGLIPLSSFAVNVVVALTLGAGTDYGIFLLGRYHEA RQSGESREDAYFTAYRSVAPIIIGSGLTIAGACFCLSLARLDYFHTMGPAVAISMLFTIAAALTLAPALL TLGSLLALFDPKRPTRGHLYRRIGTSVVRWPKPIFVASAAVVMVGAVFVPSYRVNYDDRAYQPVDAAANL GFQAADRHFNQSKLFSEMLMIETDHDMRNSADFISLDRVAKALIRLPGVAMVQSITRPLGRPLEHATIPY LFTTQGSTNGQQLPFSEQANENTDRQAEIQARSVEVLQHVITITQQMSAELHSTVLTFENLKEVTDEVNE GISNLDDFLRPLKNYFYWEPHCFDIPVCFAMRSLFDSLDGIDSLDEQIGNAVVSFEAIDRLLPQMITQLE RMIADSRALLGVITNNYGPAHLQSTQTDQTYYDQINVGNDFDAARSDDFFYIPHEAFDNDDVKTGMKLLM SPDGKAARFIITHEGNAMAPEGIEHVEQFPDAIKMALKETSLAGAKIYIGGAASNNLDIKTYAASDLLIV AIAAFVLIFLIMLYLTRSLVAALVIPGTVAFSFAGAFGLSVMMWQHLIGLPLHWLILPLSFIILVAVGSD YNLLLISRVKEETGAGLNTGLIRALGSTGAVVTSAGLVFAFTMLSMLVSDLRTIGQLGTTVCIGLLLDTM IVRSLVVPCLLRLLGPWFWWPTFIRQRPLRQAVRQRERADVV*
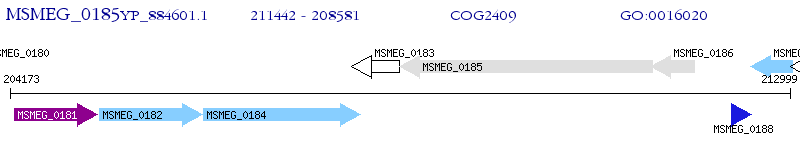
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0185 | - | - | 100% (953) | MmpL6 protein |
| M. smegmatis MC2 155 | MSMEG_3584 | - | 0.0 | 84.59% (941) | membrane protein, MmpL family protein |
| M. smegmatis MC2 155 | MSMEG_0576 | - | 0.0 | 47.89% (925) | MmpL4 protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1583 | mmpL6 | 0.0 | 48.14% (939) | transmembrane transport protein MmpL6 |
| M. gilvum PYR-GCK | Mflv_4418 | - | 0.0 | 47.29% (941) | transport protein |
| M. tuberculosis H37Rv | Rv0450c | mmpL4 | 0.0 | 48.44% (931) | transmembrane transport protein MmpL4 |
| M. leprae Br4923 | MLBr_02378 | - | 0.0 | 48.06% (926) | hypothetical protein MLBr_02378 |
| M. abscessus ATCC 19977 | MAB_4768c | - | 0.0 | 75.16% (938) | putative membrane protein, MmpL |
| M. marinum M | MMAR_3744 | mmpL4_1 | 0.0 | 48.65% (929) | transmembrane transport protein MmpL4_1 |
| M. avium 104 | MAV_3863 | - | 0.0 | 48.24% (939) | MmpL4 protein |
| M. thermoresistible (build 8) | TH_3260 | - | 0.0 | 44.21% (941) | PUTATIVE MmpL4 protein |
| M. ulcerans Agy99 | MUL_3688 | mmpL4_1 | 0.0 | 48.44% (929) | transmembrane transport protein MmpL4_1 |
| M. vanbaalenii PYR-1 | Mvan_3189 | - | 0.0 | 46.15% (934) | transport protein |
CLUSTAL 2.0.9 multiple sequence alignment
Rv0450c|M.tuberculosis_H37Rv ---MSTKFANDSNTNARPEK------------------PFIARMIHAFAV
MLBr_02378|M.leprae_Br4923 ---MTVKCANDLDTHTKP--------------------PFVARMIHRFAV
TH_3260|M.thermoresistible__bu ---MTANPGHVRTDEAPTDRLDI--------KVSTHGPSFISTWIRRLSL
Mvan_3189|M.vanbaalenii_PYR-1 ---MSAPVSDARTDEFPAAR-------------QPHRP-FIPRMIRLFAV
Mflv_4418|M.gilvum_PYR-GCK ----MADAGRSG----------------------------IGRIIRVLAV
Mb1583|M.bovis_AF2122/97 ---MSNHHRPRP---------------------------WLPHTIRRLSL
MAV_3863|M.avium_104 ---MTTDQATRS---------------------------VVAQTIRRLSV
MMAR_3744|M.marinum_M ---MSESNSGASISTEDTGPIEIGAATGGAPNRERGHRPVIPHTIRILAV
MUL_3688|M.ulcerans_Agy99 ---MSESNSGASISTEDTGPIEIGAATGGAPNRERGHRPVIPHTIRILAV
MSMEG_0185|M.smegmatis_MC2_155 MPGQVRMSAHRAGR------------------------PFFARTVRVLAV
MAB_4768c|M.abscessus_ATCC_199 ---MSEQSKHSVKR------------------------PLIPRMVRVLAV
. :: :::
Rv0450c|M.tuberculosis_H37Rv PIILGWLAVCVVVTVFVPSLEAVGQERSVSLSPKDAPSFEAMGRIGMVFK
MLBr_02378|M.leprae_Br4923 PIILVWLAIAVTVSVFIPSLEDVGQERSVSLSPKDAPSYIAMQKMGQVFN
TH_3260|M.thermoresistible__bu PIIVGWIALIALLNITVPQLEVVGEMRAVSMSPDEAPSMVAMKRVGELFD
Mvan_3189|M.vanbaalenii_PYR-1 PIILGWIALIVILNVTVPQLEAVGEARAVSMSPNEAPSLISMKKVGELFR
Mflv_4418|M.gilvum_PYR-GCK PIVLGWVLLTVLTNVAVPSLEKVGEEHTVGMSAGDAPSMMSMKRVGANFE
Mb1583|M.bovis_AF2122/97 PILLFWVGVAAITNAAVPQLEVVGEAHNVAQSSPDDPSLQAMKRIGKVFH
MAV_3863|M.avium_104 PILLVWVAVAAISNIAVPNLEDVAKAHNVSLNTPAAPSFQAMQHIGKVFH
MMAR_3744|M.marinum_M PIILGWVLLTVLVNVVVPSLEVVSEQHSAPMTPMDAPSMKAMMRLGHNFR
MUL_3688|M.ulcerans_Agy99 PIILGWVLLTVLVNVVVPSLEVVSEQHSAPMTPMDAPSMKAMMRLGHNFR
MSMEG_0185|M.smegmatis_MC2_155 PIIVFWGLLAVTTNTFIPQVEHVADELAGPMIPNYAPSQVALLHIGEKFQ
MAB_4768c|M.abscessus_ATCC_199 PIVVFWAIVAVTTNTFVPHVEDVAEELAGPMIPHYAPSQRAMLHIGEKFQ
**:: * : . . :* :* *.. . ** :: ::* *
Rv0450c|M.tuberculosis_H37Rv EGDSDSFAMVIIEGNQPLGDAAHKYYDGLVAQLRADKKHVQSVQDLWGDP
MLBr_02378|M.leprae_Br4923 EGNSDSVIMIVLEGDKPLGDDAHRFYDGLIRKLRADK-HVQSVQDFWGDP
TH_3260|M.thermoresistible__bu EGSSDSQVMIVLEGEEPLGADARAFYDELVKRLEADDAHVENVQDLWSDP
Mvan_3189|M.vanbaalenii_PYR-1 EGDSDSSVMIVFEGDQPLGDEAHAWYDELVERLRADTKHVQSVQDFWSDP
Mflv_4418|M.gilvum_PYR-GCK EFDSDSSAMVVLEGEQPLGDAAHRYYDQLIDELEADTANVQHIADFWGDP
Mb1583|M.bovis_AF2122/97 EFDSDSAAMIVLEGDKPLGNDAHRFYDTLLRNLSNDTKHVEHVQDFWGDP
MAV_3863|M.avium_104 EYDSDSAAMIVLEGDKPLGDDAHRFYDALVGKLEHDTKHVEHVQNFWGDP
MMAR_3744|M.marinum_M EFDSNSTVMIVLEGQHKLGDDAHQYYDSLIRQLRQDPEHIQHIQDFWGDR
MUL_3688|M.ulcerans_Agy99 EFDSNSTVMIVLEGQHKLGDDAHQYYDSLIRQLRQDPEHIQHIQDFWGDR
MSMEG_0185|M.smegmatis_MC2_155 ESDSTNLTMLVFEADRPLDDGDHRYYDDLVRRLQADTEHVQYVMDLWGKP
MAB_4768c|M.abscessus_ATCC_199 ESTSTSLTMLVLEADRPLDGSDHQYYDDLVRRLKSDTEHVQYVMDTWGKP
* * . *:::*.:. *. : :** *: .* * ::: : : *..
Rv0450c|M.tuberculosis_H37Rv LTAAGVQSNDGKAAYVQLSLAGNQGTPLANESVEAVRSIVESTP---APP
MLBr_02378|M.leprae_Br4923 LTAPGAQSNDGKAAYVQLSLAGNQGELLAQESIDAVRKIVVQTP---APP
TH_3260|M.thermoresistible__bu LTATGVQSPDGKSAYVQVKLAGNQGEALANESVETVREMIATVSEEMAPE
Mvan_3189|M.vanbaalenii_PYR-1 LTASGSQSNDGKAAYVQVKLAGNQGESLANESVQAAQEIVRSLE---PPP
Mflv_4418|M.gilvum_PYR-GCK LTASGAQSTDGKAAYVQVYLQGNQGEPRANEAVAAVREIVDRTP---APP
Mb1583|M.bovis_AF2122/97 LTAAGSQSTDGKAAYVQVYLAGNQGEALSIESVDAVRDIVAHTP---PPA
MAV_3863|M.avium_104 LTAAGSQSKDGKAAYVQVYLAGNQGDALANESVDAIRNIVEHTP---PPP
MMAR_3744|M.marinum_M LTAAGAQSADGKSAYVMVNLAGNQGTTLANESVEAVRDVINRTH---EPA
MUL_3688|M.ulcerans_Agy99 LTAAGAQSADGKSAYVMVNLAGNQGTTLANESVEAVRDVINRTH---EPA
MSMEG_0185|M.smegmatis_MC2_155 ITAAGAQSVDGKASFVLLRLAGNIGQLEANESVDAVRDIIADDA---PPQ
MAB_4768c|M.abscessus_ATCC_199 ITAAGAQSLDGKSAFVLLRLAGDIGQTQANKSVEAIKETIDKDT---PPR
:**.* ** ***:::* : * *: * : ::: : :. : *
Rv0450c|M.tuberculosis_H37Rv GIKAYVTGPSALAADMHHSGDRSMARITMVTVAVIFIMLLLVYRSIITVV
MLBr_02378|M.leprae_Br4923 GITAWVTGASALIADMHHSGDKSMIRITATSVIVILVTLLLVYRSFITVI
TH_3260|M.thermoresistible__bu GVQAFVTGGAAMSADQQEAGDSSLRVIEALTFLVITTMLLFVYRSVITVL
Mvan_3189|M.vanbaalenii_PYR-1 GVRAFVTGPAALAADQHIASDRSVRVIELVTFAVIIVMLLLVYRSIVTVL
Mflv_4418|M.gilvum_PYR-GCK GIKAYVTGGAPLVADQHSAGDTSVLRVTLITIGVIAVMLLIVFRSVATMV
Mb1583|M.bovis_AF2122/97 GVKAYVTGAAPLMADQFQVGSKGTAKVTGITLVVIAVMLLFVYRSVVTMV
MAV_3863|M.avium_104 GVKAYVTGAAPLVTDQFEVGSKGIFKVTVITVLVILAMLLWVYRS-VTAV
MMAR_3744|M.marinum_M GVKAYVTGPAALSDDMHLIGNASLAKITMFTLGAIAVMLLLVYRSIVTTV
MUL_3688|M.ulcerans_Agy99 GVKAYVTGPAALSDDMHLIGNASLATITMFTLGAIAVMLLLVYRSIVTTV
MSMEG_0185|M.smegmatis_MC2_155 GLKVYVSGAAPLASDTLSTANSSLNNITIFTIILIVVMLLLVYRSAACLL
MAB_4768c|M.abscessus_ATCC_199 GLKVYVSGAAPLASDTLSVANSSLNNITIVTIFLIIFMLLMVYRSVTTML
*: .:*:* :.: * .. . : :. * ** *:** :
Rv0450c|M.tuberculosis_H37Rv LLLITVGVELTAARGVVAVLGHSGAIGLTTFAVSLLTSLAIAAGTDYGIF
MLBr_02378|M.leprae_Br4923 LLLFTVGIESAVARGVVALLGHTGLIGLSTFAVNLLTSLAIAAGTDYGIF
TH_3260|M.thermoresistible__bu IALFMVLIGLLTTRGVVAFLGYHNIIGLSTFATSLLVTLAIAIAVDYAIF
Mvan_3189|M.vanbaalenii_PYR-1 LTLVMVVLSLATARGVVAFLGWHEIIGLSLFATNLLVTLAIAAATDYAIF
Mflv_4418|M.gilvum_PYR-GCK LILFMVFAELGAARGIVAFLADAEIIGLSTFATSLLTLMVIAAGTDYAIF
Mb1583|M.bovis_AF2122/97 LVLITVLIELAAARGIVAFLGNAGVIGLSTYSTNLLTLLVIAAGTDYAIF
MAV_3863|M.avium_104 FVLVTVIVEMAAARGIVAFLGSVGLIGLSTYATNLLTLLVIAAGTDYAIF
MMAR_3744|M.marinum_M IQLFMTGIALASSRGVVAVLGYHNVFGLTTFAANILTMLAIAAGTDYGIF
MUL_3688|M.ulcerans_Agy99 IQLFMTGIALASSRGVVAVLGYHNVFGLTTFAANILTMLAIAAGTDYGIF
MSMEG_0185|M.smegmatis_MC2_155 VPLPAVLIEMLVAKGVIATLGHFGLIPLSSFAVNVVVALTLGAGTDYGIF
MAB_4768c|M.abscessus_ATCC_199 VPLFGVLVEMLVARGVVATLGHFGYIELSSFAVNIVIALTLGAGTDYGIF
. * . ::*::* *. : *: ::..:: :.:. ..**.**
Rv0450c|M.tuberculosis_H37Rv IIGRYQEARQAGEDKEAAYYTMYRGTAHVILGSGLTIAGATFCLSFARMP
MLBr_02378|M.leprae_Br4923 ITGRYQEARQANENKEAAFYTMYRGTFHVILGSGLTISGATFCLSFARMP
TH_3260|M.thermoresistible__bu LIGRYQEARSVGADRATAYHTMFGGTAHVILASGLTIAGATFCLSFTRLP
Mvan_3189|M.vanbaalenii_PYR-1 LIGRYQEARTNGEDKESAYYTMFHGTAHVVLGSGLTIAGATYCLSFTRLP
Mflv_4418|M.gilvum_PYR-GCK AIGRYQEARGAGEDRETAYYTMFRGTAHVVLGSGLTIAGAMLCLSFTRLP
Mb1583|M.bovis_AF2122/97 VLGRYHEARYAAQDRETAFYTMYRGTAHVVLGSGLTVAGAVYCLSFTRLP
MAV_3863|M.avium_104 FVGRYHEARHEGQDRETAYYTMYRGTTHVVLGSGLTVAGAVLCLRFTRLN
MMAR_3744|M.marinum_M LVGRYQEARQAGEDREDAYYTTFRGVAPVVLGSGLTIAGATYCLSFARLP
MUL_3688|M.ulcerans_Agy99 LVGRYQEARQAGEDRQDAYYTTFRGVAPVVLGSGLTIAGATYCLSFARLP
MSMEG_0185|M.smegmatis_MC2_155 LLGRYHEARQSGESREDAYFTAYRSVAPIIIGSGLTIAGACFCLSLARLD
MAB_4768c|M.abscessus_ATCC_199 LLGRYHEARNAGLDREDAFYSAYRGVSHVIIGSGLTIAGAGLCLSFARLN
***:*** .: *:.: : .. :::.****::** ** ::*:
Rv0450c|M.tuberculosis_H37Rv YFQTLGIPCAVGMLVAVAVALTLGPAVLHVGSRFG-LFDPKRLLKV-RGW
MLBr_02378|M.leprae_Br4923 YFQTLGVPCAVGMLIAVAVALTLGPAVLTVGSRFG-LFEPKRLIKV-RGW
TH_3260|M.thermoresistible__bu YFQSLGPPLAIGMIVLIVTSLTMGPAIITVATRFG-LLEPKRAARV-RFW
Mvan_3189|M.vanbaalenii_PYR-1 YFQTLGVPLAIGMFVVVMAGVILMVAMISVATRFGKLLEPKRAMRI-RGW
Mflv_4418|M.gilvum_PYR-GCK YFQTMGVPCAVGTFVAVIAALTLGPAVIVIGSRFG-LFDPKRAISS-RGW
Mb1583|M.bovis_AF2122/97 YFQSLGIPASIGVMIALAAALSLAPSVLILGSRFG-CFEPKRRMRT-RGW
MAV_3863|M.avium_104 YFQSLGIPAAIGIGVALAAALSLTPAVITVGSLFG-LFDPKRQMAT-RGW
MMAR_3744|M.marinum_M WFNTMGAPVAIGMLVVVLAGLTLGPAVVFVGSRFH-LFESKRAANRGRLW
MUL_3688|M.ulcerans_Agy99 WFNTMGAPVAIGMLVVVLAGLTLGPAVVFVGSRFH-PFESKRAANRGRLW
MSMEG_0185|M.smegmatis_MC2_155 YFHTMGPAVAISMLFTIAAALTLAPALLTLGSLLA-LFDPKRPTRG-HLY
MAB_4768c|M.abscessus_ATCC_199 YFHTMGPAVAIAMLLTIAAALTLGPALLHLGSLFG-LFDPKRRVKA-RLY
:*:::* . ::. . : ..: : ::: :.: : ::.** : :
Rv0450c|M.tuberculosis_H37Rv RRVGTVVVRWPLPVLVATCAIALVGLLALPGYKTSYNDRDYLPDFIPANQ
MLBr_02378|M.leprae_Br4923 RRIGTVVVRWPLPILITTCAIAMVGLLALPGYRTNYKDRAYLPASIPANQ
TH_3260|M.thermoresistible__bu RKVGASVVRWPGPIMVTTVAVTLVGLLALPGYRTNYNDRNYLPASFPSNA
Mvan_3189|M.vanbaalenii_PYR-1 RKIGAAVVRWPGPILVATLAITLVGLLALPGYKTNYNDRTYLPADLPANE
Mflv_4418|M.gilvum_PYR-GCK RRIGVSVVRWPGPVLAATLALALVGLVTLPGYKTNYDARDYLPEDIPANV
Mb1583|M.bovis_AF2122/97 RRIGTAIVRWPGPILAVACAIAVVGLLALPGYKTSYDARYYMPATAPANI
MAV_3863|M.avium_104 RRIGTAIVRWPGPILVVATGVALVGLLALPGYRTNYDARPYMPASAPANT
MMAR_3744|M.marinum_M RRVGTAVVRWPAPILALSGAIVLVGMVALPGFKTSYNDRYYLPASAPSNL
MUL_3688|M.ulcerans_Agy99 RRVGTAVVRWPAPILALSGAIVLVGMVALPGFKTSYNDRYYLPASAPSNL
MSMEG_0185|M.smegmatis_MC2_155 RRIGTSVVRWPKPIFVASAAVVMVGAVFVPSYRVNYDDRAYQPVDAAANL
MAB_4768c|M.abscessus_ATCC_199 RRIGASVVRWPKPILVASAAAVLVGAVFVPTYQVNYDDRAYQPPDAPAKQ
*::*. :**** *:: : . .:** : :* ::..*. * * * .::
Rv0450c|M.tuberculosis_H37Rv GYAAADRHFSQARMKPEILMIESDHDMRNPADFLVLDKLAKGIFRVPGIS
MLBr_02378|M.leprae_Br4923 GFAAADRHFPQARMKPEILMIESDHDMRNPADFLILDKLARGIFRVPGIS
TH_3260|M.thermoresistible__bu GYEAAERHFDPARLNPELLLIESDHDLRNPADFLVIEKIAKAVFQVEGIS
Mvan_3189|M.vanbaalenii_PYR-1 GYAVADRHFDQARMNPELLMIESDHDLRNSADFLVIDKIAKAIFKVEGIA
Mflv_4418|M.gilvum_PYR-GCK GYAVADQHFGSARMNPELLMVESDHDLRNPADFLVIDKIAKSVFRVPGVA
Mb1583|M.bovis_AF2122/97 GYMAAERHFPQARLNPELLMIETDHDMRNPADMLILDRIAKAVFHLPGIG
MAV_3863|M.avium_104 GYTAAERHFSQARLNPELLMIETDHDMRNPADMINLERVAKAIAHLPGIA
MMAR_3744|M.marinum_M GEAAADRHFSHARMNPDMLMVEAEHDMRNPADMLVLDKVAKNVIRTVGIA
MUL_3688|M.ulcerans_Agy99 GEAAADRHFSHARMNPDMLMVEAEHDMRNPADMLVLDKVAKNVIRTVGIA
MSMEG_0185|M.smegmatis_MC2_155 GFQAADRHFNQSKLFSEMLMIETDHDMRNSADFISLDRVAKALIRLPGVA
MAB_4768c|M.abscessus_ATCC_199 GFAAADRHFPPSKLFTEMLMVESDHDMRNSADFISLDRVSKSLVRLPGVA
* .*::** ::: .::*::*::**:**.**:: :::::: : : *:.
Rv0450c|M.tuberculosis_H37Rv RVQAITRPEGTTMDHTSIPFQISMQNAGQLQTIKYQRDRANDMLKQADEM
MLBr_02378|M.leprae_Br4923 RVQAITRPDGTAMDHTSIPFQISMQNAGQVQTMKYQKDRMNDLLRQAENM
TH_3260|M.thermoresistible__bu RVQGITRPEGTPIEHSSIPFMISQQGTLQILNQQYMQDRMADMLVQADEM
Mvan_3189|M.vanbaalenii_PYR-1 RVQAITRPDGKPIKHTSIPFQMSMQGTTQRLNEKYMQDRMADMLVQADEM
Mflv_4418|M.gilvum_PYR-GCK RVQTITRPDGKPIKHTTIPFQMSMQGTTQRLNEKYLQDRMADMLVQADAM
Mb1583|M.bovis_AF2122/97 LVQAMTRPLGTPIDHSSIPFQISMQSVGQIQNLKYQRDRAADLLKQAEEL
MAV_3863|M.avium_104 QVQSMTRPLGTPIEHTSLAFQISAGSIGSIENLQYQKERAEDLLKQADNL
MMAR_3744|M.marinum_M MIQDITRPLGIPIQHSSIPFQNSMQSQTMMQNMAFLKERMADMTKMADEM
MUL_3688|M.ulcerans_Agy99 MIQDITRPLGIPIQHSSIPFQNSMQSQTMMQNMAFLKERMADMTKMADEM
MSMEG_0185|M.smegmatis_MC2_155 MVQSITRPLGRPLEHATIPYLFTTQGSTNGQQLPFSEQANENTDRQAEIQ
MAB_4768c|M.abscessus_ATCC_199 MVQSITRPMGRALEHASLPYLFTTQGSGNGQQLPFTKAQNANTDRQAQIM
:* :*** * .:.*:::.: : . : . : *:
Rv0450c|M.tuberculosis_H37Rv ATTIAVLTRMHSLMAEMASTTHRMVGDTEEMKEITEELRDHVADFDDFWR
MLBr_02378|M.leprae_Br4923 AETIASMRRMHQLMALLTENTHHILNDTVEMQKTTSKLRDEIANFDDFWR
TH_3260|M.thermoresistible__bu QLMIDTMDKMIALMEEMVETTDNMVRDMDEMAAHVAEMRDSISEFDDFFR
Mvan_3189|M.vanbaalenii_PYR-1 ANTIATMEKMSNLTAQMADITHSMVSKMENMLTDIEDLRDSIANFDDFFR
Mflv_4418|M.gilvum_PYR-GCK QTNVDTMTRMQSLMGQMSATTHEMVLKTKKTEIDIIEVRDNLADFDDFFR
Mb1583|M.bovis_AF2122/97 GKTIEILQRQYALQQELAAATHEQAESFHQTIATVKELRDRIANFDDFFR
MAV_3863|M.avium_104 KDTIGILNQQYALQKQLAASTHDETQSFHDTIAIINDLRDKIANFDDFFR
MMAR_3744|M.marinum_M QVLIDTTERMYAVMRDLDTTTTDMARVTAETSTITDELRDHLAEFDDFWR
MUL_3688|M.ulcerans_Agy99 QVLIDTTERMYAVMRELDTTTTDMARVTAETSTITDELRDHLAEFDDFWR
MSMEG_0185|M.smegmatis_MC2_155 ARSVEVLQHVITITQQMSAELHSTVLTFENLKEVTDEVNEGISNLDDFLR
MAB_4768c|M.abscessus_ATCC_199 AHSVAVLRQTTDLTQQLADEMHNTVVTMEDMQQVTQDMNEKVSNLDDFMR
: : : : . .:.: ::::*** *
Rv0450c|M.tuberculosis_H37Rv PIRSYFYWEKHCYGIPICWSFRSIFDALDGIDKLSEQIGVLLGDLREMDR
MLBr_02378|M.leprae_Br4923 PIRSYFYWERHCFNIPICWSFRSIFDALDGVDQIDERLNSIVGDIKNMDL
TH_3260|M.thermoresistible__bu PIRNYFYWEPHCYNIPVCWAMRSVFDTLDGINTMTDDIQNMLPHMHRLND
Mvan_3189|M.vanbaalenii_PYR-1 PIRNYFYWEPHCYNIPVCWALRSVFDTLDGINVMTDDFREILPDMKRLDS
Mflv_4418|M.gilvum_PYR-GCK PVRNYFYWEPHCYDIPVCWTMRSVFDTLDGINPLTDDISELIPDLERLDE
Mb1583|M.bovis_AF2122/97 PIRSYFYWEKHCYDIPSCWALRSVFDTIDGIDQLGEQLASVTVTLDKLAA
MAV_3863|M.avium_104 PVRSYFYWEKHCFDIPACFAFRSVFDALDGIDELSAQFEKLTASLDKLDA
MMAR_3744|M.marinum_M PMRSYFYWEKHCFDVPICWSFRSLFDSLDGIDKLSEKFSELTEDIQHTAG
MUL_3688|M.ulcerans_Agy99 PMRSYFYWEKHCFDVPICWSFRSLFDSLDGIDKLSEKFSELTEDIQRTAG
MSMEG_0185|M.smegmatis_MC2_155 PLKNYFYWEPHCFDIPVCFAMRSLFDSLDGIDSLDEQIGNAVVSFEAIDR
MAB_4768c|M.abscessus_ATCC_199 PLRNYFYWEPHCFDIPVCQSFRSLFDMLDSIDKLAADIKDAVGSLEVVDR
*::.***** **:.:* * ::**:** :*.:: : : :
Rv0450c|M.tuberculosis_H37Rv LMPQMVAQIPPQIEAMENMRTMILTMHSTMTGIFDQMLEMSDNATAMGKA
MLBr_02378|M.leprae_Br4923 LMPQMLEQFPPMIESMESMRTIMLTMHSTMSGIFDQMNELSDNANTMGKA
TH_3260|M.thermoresistible__bu LMPQMLTLLPTTVATMKTMRSMLLTMQATNAGLQDQMAEMQENANAMGEA
Mvan_3189|M.vanbaalenii_PYR-1 LMPQMVALMPEMIETMKTMRTMMLTMYQSQKGQQDQMAAMSEDADAMGEA
Mflv_4418|M.gilvum_PYR-GCK LMPQLIALLPSQIETMRSMQAMMLRQYQTQKGQLDQNAAMSEDADAMGEA
Mb1583|M.bovis_AF2122/97 IQPQLVALLPDEIASQQINRELALANYATMSGIYAQTAALIENAAAMGQA
MAV_3863|M.avium_104 GQQKLVTLLPPQIADQEKNLALTLSNYATNLGINAQTRANTDTATALGQA
MMAR_3744|M.marinum_M LTSQMLTLIPPMIATMKTTKALTLTMQSTFSAMLDQMDELSNTAIVMGQS
MUL_3688|M.ulcerans_Agy99 LTSQMLTLIPPMIATMKTTKALTLTMQSTFSAMLDQMDELSNTAIVMGQS
MSMEG_0185|M.smegmatis_MC2_155 LLPQMITQLERMIADSRALLGVITNNYGPAHLQSTQTDQTYYDQINVGND
MAB_4768c|M.abscessus_ATCC_199 LLPQMVALLKLTANDQEALQALIVNTYGQTNLQATATDQTFDDMINVGND
::: : . : :*:
Rv0450c|M.tuberculosis_H37Rv FDAAKNDDSFYLPPEVFKNKDFQRAMKSFLSSDGHAARFIILHRGDPQSP
MLBr_02378|M.leprae_Br4923 FDTAKNDDSFYLPPEVFKNTDFKRAMKSFLSSDGHAARFIILHRGDPASV
TH_3260|M.thermoresistible__bu FDQARNDDSFYLPPEAFDNPDFQRGLEQFLSPDGHAVRFIISHQGDPMTP
Mvan_3189|M.vanbaalenii_PYR-1 FDDSMNDDSFYLPPEIFENKDFQRGLEQFLSPDGHAVRFIISHEGDPLSP
Mflv_4418|M.gilvum_PYR-GCK FDDSMNDDSFYLPPEAFDNDDFKRGMENFISPDGRSVRFIISHEGDPATV
Mb1583|M.bovis_AF2122/97 FDAAKNDDSFYLPPEAFDNPDFQRGLKLFLSADGKAARMIISHEGDPATP
MAV_3863|M.avium_104 YDAAKNDDSFYLPPEAFTNPEFKRGLKLFLSPDGKAARMIISHEGDPATP
MMAR_3744|M.marinum_M FDASKNDDFFYLPPEAFDNPDFQTGLRMFLSPDGKSARFFITHQGDPMTP
MUL_3688|M.ulcerans_Agy99 FDASKNDDFFYLPPEAFDNPDFQTGLRMFLSPDGKSARFFITHQGDPMTP
MSMEG_0185|M.smegmatis_MC2_155 FDAARSDDFFYIPHEAFDNDDVKTGMKLLMSPDGKAARFIITHEGNAMAP
MAB_4768c|M.abscessus_ATCC_199 FDASRSDDFFYIPHEAFDNEDIKTGMTLMMSPDGKAARFIVTHMGNAMGP
:* : .** **:* * * * :.: .: ::*.**::.*::: * *:.
Rv0450c|M.tuberculosis_H37Rv EGIKSIDAIRTAAEESLKGTPLEDAKIYLAGTAAVFHDISEGAQWDLLIA
MLBr_02378|M.leprae_Br4923 AGIASINAIRTAAEEALKGTPLEDTKIYLAGTAAVFKDIDEGANWDLVIA
TH_3260|M.thermoresistible__bu EGLAKIEPIKKAAVEAIKGTPLEGSRVYLGGTAAVFKDMSDGTKYDLIIA
Mvan_3189|M.vanbaalenii_PYR-1 EGVAKIDKIKTAAKEAIKGTPLEGSKIYLGGTAATFKDMQDGNNYDLLIA
Mflv_4418|M.gilvum_PYR-GCK EGISEVVPIKTAAKEAIKGTPLEGSTVYLAGTAATYKDMSDGAFYDLLIA
Mb1583|M.bovis_AF2122/97 EGISHIDAIKQAAHEAVKGTPMAGAGIYLAGTAATFKDIQDGATYDLLIA
MAV_3863|M.avium_104 EGISHIEPIRNAAKQAVKNTPLGDSNIYLAGTAATYKDIAEGAKYDLLIA
MMAR_3744|M.marinum_M EGISRVDAERTAAQEGLKQSSLSDAKVYLGGTAATFKDMADGEKYDLMIA
MUL_3688|M.ulcerans_Agy99 EGISRVDAERTAAQEGLKQSSLSDAKVYLGGTAATFKDMADGEKYDLMIA
MSMEG_0185|M.smegmatis_MC2_155 EGIEHVEQFPDAIKMALKETSLAGAKIYIGGAASNNLDIKTYAASDLLIV
MAB_4768c|M.abscessus_ATCC_199 EGIEHVNDFPDAITTALKETSLAGAQVYIGGAGSNNKDIKQYAASDLLIA
*: : * .:* :.: .: :*:.*:.: *: **:*.
Rv0450c|M.tuberculosis_H37Rv AISSLCLIFIIMLIITRAFIAAAVIVGTVALSLGASFGLSVLLWQHILAI
MLBr_02378|M.leprae_Br4923 GISSLCLIFIIMLIITRAFVAAAVIVGTVALSLGASFGLSVLLWQHILGI
TH_3260|M.thermoresistible__bu AIAAVCLIFIIMLILTRAVVAAAVIVGTVTLSLAASFGVSVLIWQYIVGL
Mvan_3189|M.vanbaalenii_PYR-1 GISALGLIFIIMLILTRAIVAAAVIVGTVVLSLAASFGLSVLFWQHILGT
Mflv_4418|M.gilvum_PYR-GCK GIAAVTLIFVIMLVITRSVVAAAVIVGTVLLSLGASFGLSVLLWQHIIGL
Mb1583|M.bovis_AF2122/97 GIAALSLILLIMMIITRSLVAALVIVGTVALSLGASFGLSVLVWQHLLGI
MAV_3863|M.avium_104 GIAALSLILLIMVLITRSLVAAIVIVGTVALSLGASFGLSVLVWQDILGI
MMAR_3744|M.marinum_M VVSALTLIFLIMLLLTRSVVAALVIVGTAASSIAASFGLSVLIWQDLFGI
MUL_3688|M.ulcerans_Agy99 VVSALTLIFLIMLLLTRSVVAALVIVGTAASSIAASFGLSVLIWQDLFGI
MSMEG_0185|M.smegmatis_MC2_155 AIAAFVLIFLIMLYLTRSLVAALVIPGTVAFSFAGAFGLSVMMWQHLIGL
MAB_4768c|M.abscessus_ATCC_199 AIAAFTLIFLIMMVITRSLVAPFVIIGTVAFSFAGAFGLSVLIWQHLVGL
:::. **::**: :**:.:*. ** **. *:..:**:**:.** :..
Rv0450c|M.tuberculosis_H37Rv HLHWLVLAMSVIVLLAVGSDYNLLLVSRFKQEIGAGLKTGIIRSMGGTGK
MLBr_02378|M.leprae_Br4923 ELHYLVLAMSVIVLLAVGSDYNLLLVSRFKQEIQAGLKTGIIRSMGGTGK
TH_3260|M.thermoresistible__bu EMHWMVFPMALIILLAVGADYNLLVVARLREEIPAGLNTGMIRAMGGSGS
Mvan_3189|M.vanbaalenii_PYR-1 ELHWMVLAMAVIVLLAVGADYNLLLVSRLKEEIHAGIGTGIIRAMGGSGS
Mflv_4418|M.gilvum_PYR-GCK ALHWMVLPMAVILLLAVGSDYNLLLVSRFKEELSGGLKTGIIRAMAGTGS
Mb1583|M.bovis_AF2122/97 QLYWIVLALAVILLLAVGSDYNLLLISRFKEEIGAGLNTGIIRAMAGTGG
MAV_3863|M.avium_104 KLYWICLALSVIILLAVGSDYNLLLISRFREEIHAGLNTGIIRSMAGSGA
MMAR_3744|M.marinum_M KIHWIVAALSVIILLAVGSDYNLLLVSRFREEIHAGLKTGIIRSMAGTGG
MUL_3688|M.ulcerans_Agy99 KIHWIVAALSVIILLAVGSDYNLLLVSRFREEIHAGLKTGIIRSMAGTGG
MSMEG_0185|M.smegmatis_MC2_155 PLHWLILPLSFIILVAVGSDYNLLLISRVKEETGAGLNTGLIRALGSTGA
MAB_4768c|M.abscessus_ATCC_199 PLHWLILPLTFIILVAVGSDYNLLLIARLKEETDAGLNTGLIRALGSTGG
:::: .::.*:*:***:*****:::*.::* .*: **:**::..:*
Rv0450c|M.tuberculosis_H37Rv VVTNAGLVFAVTMASMAVSDLRVIGQVGTTIGLGLLFDTLIVRSFMTPSI
MLBr_02378|M.leprae_Br4923 VVTNAGLVFAFTMASMVVSDLRVIGQVGTTIGLGLLFDTLIVRSFMMPSI
TH_3260|M.thermoresistible__bu VVTAAGLVFGLTMMVMVVSDLTVMGQVGTTIGLGLLFDTFVVRSFMMPSI
Mvan_3189|M.vanbaalenii_PYR-1 VVTAAGLVFALTMMSMAVSELTVIGQVGTTIGLGLLFDTLVIRAFMTPSI
Mflv_4418|M.gilvum_PYR-GCK VVTSAGLVFAFTMASFAFSDLKVMAQVGTTIALGLLFDTLIVRSFMTPAV
Mb1583|M.bovis_AF2122/97 VVTAAGLVFAATMSSFVFSDLRVLGQIGTTIGLGLLFDTLVVRAFMTPSI
MAV_3863|M.avium_104 VVTSAGLVFAFTMASFVSASLLVLGQIGTTIALGLLFDTLIVRSFMTPSV
MMAR_3744|M.marinum_M VVTAAGLVFAFTMASMLGSDLRVLGQFGSTVCIGLLLDTLIVRTLLMPSI
MUL_3688|M.ulcerans_Agy99 VVTAAGLVFAFTMASMLGSDLRVLGQFGSTVCIGLLLDTLIVRTLLMPSI
MSMEG_0185|M.smegmatis_MC2_155 VVTSAGLVFAFTMLSMLVSDLRTIGQLGTTVCIGLLLDTMIVRSLVVPCL
MAB_4768c|M.abscessus_ATCC_199 VVTSAGLVFAFTMLAMLASDLRTIGQVGTTVCIGLLLDTLIVRSFIVPAL
*** *****. ** : :.* .:.*.*:*: :***:**:::*::: *.:
Rv0450c|M.tuberculosis_H37Rv AALLGRWFWWPLRVRSRPARTPTVPSETQPAGRPLAMSSDRLG-------
MLBr_02378|M.leprae_Br4923 AALLGRWFWWPQQGRTRPLLTVAAPVGLPPD-RSLVYSD-----------
TH_3260|M.thermoresistible__bu ATLLGRWFWWPQFVRKRPVPQPWPSVPPRGPATTDRVSV-----------
Mvan_3189|M.vanbaalenii_PYR-1 AALLGPWFWWPQRVRTRPVPAPWPRPGGLQSDPSEGVKV-----------
Mflv_4418|M.gilvum_PYR-GCK AALLGRWFWWPQKFRSAASERRLARIG-VQPATAGR--------------
Mb1583|M.bovis_AF2122/97 AVLLGRWFWWPQRVRPRPASRMLRPYGPRPVVRELLLREGND---DPRTQ
MAV_3863|M.avium_104 AALLGRWFWWPQRVRPRPASTMLRPYGPRPAVRQLLLWEDGDPAVAPETP
MMAR_3744|M.marinum_M ATLLGRWFWWPQVVHPRGDNARPRVAAPTH--------------------
MUL_3688|M.ulcerans_Agy99 ATLLGRWFWWPQVVHPRGDNARPRVAAPTH--------------------
MSMEG_0185|M.smegmatis_MC2_155 LRLLGPWFWWPTFIRQRPLRQAVRQRERADVV------------------
MAB_4768c|M.abscessus_ATCC_199 VRLLGTWFWWPTRVLARPRREPVRS-------------------------
*** *****
Rv0450c|M.tuberculosis_H37Rv --------
MLBr_02378|M.leprae_Br4923 --------
TH_3260|M.thermoresistible__bu --------
Mvan_3189|M.vanbaalenii_PYR-1 --------
Mflv_4418|M.gilvum_PYR-GCK --------
Mb1583|M.bovis_AF2122/97 VATHR---
MAV_3863|M.avium_104 SAARHSRG
MMAR_3744|M.marinum_M --------
MUL_3688|M.ulcerans_Agy99 --------
MSMEG_0185|M.smegmatis_MC2_155 --------
MAB_4768c|M.abscessus_ATCC_199 --------