For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSAPVSDARTDEFPAARQPHRPFIPRMIRLFAVPIILGWIALIVILNVTVPQLEAVGEARAVSMSPNEAP SLISMKKVGELFREGDSDSSVMIVFEGDQPLGDEAHAWYDELVERLRADTKHVQSVQDFWSDPLTASGSQ SNDGKAAYVQVKLAGNQGESLANESVQAAQEIVRSLEPPPGVRAFVTGPAALAADQHIASDRSVRVIELV TFAVIIVMLLLVYRSIVTVLLTLVMVVLSLATARGVVAFLGWHEIIGLSLFATNLLVTLAIAAATDYAIF LIGRYQEARTNGEDKESAYYTMFHGTAHVVLGSGLTIAGATYCLSFTRLPYFQTLGVPLAIGMFVVVMAG VILMVAMISVATRFGKLLEPKRAMRIRGWRKIGAAVVRWPGPILVATLAITLVGLLALPGYKTNYNDRTY LPADLPANEGYAVADRHFDQARMNPELLMIESDHDLRNSADFLVIDKIAKAIFKVEGIARVQAITRPDGK PIKHTSIPFQMSMQGTTQRLNEKYMQDRMADMLVQADEMANTIATMEKMSNLTAQMADITHSMVSKMENM LTDIEDLRDSIANFDDFFRPIRNYFYWEPHCYNIPVCWALRSVFDTLDGINVMTDDFREILPDMKRLDSL MPQMVALMPEMIETMKTMRTMMLTMYQSQKGQQDQMAAMSEDADAMGEAFDDSMNDDSFYLPPEIFENKD FQRGLEQFLSPDGHAVRFIISHEGDPLSPEGVAKIDKIKTAAKEAIKGTPLEGSKIYLGGTAATFKDMQD GNNYDLLIAGISALGLIFIIMLILTRAIVAAAVIVGTVVLSLAASFGLSVLFWQHILGTELHWMVLAMAV IVLLAVGADYNLLLVSRLKEEIHAGIGTGIIRAMGGSGSVVTAAGLVFALTMMSMAVSELTVIGQVGTTI GLGLLFDTLVIRAFMTPSIAALLGPWFWWPQRVRTRPVPAPWPRPGGLQSDPSEGVKV
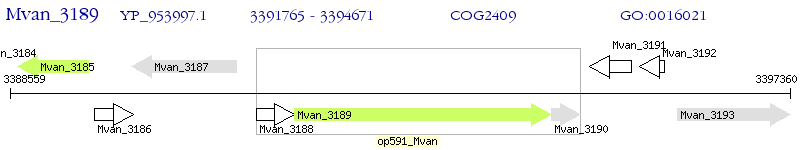
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3189 | - | - | 100% (968) | transport protein |
| M. vanbaalenii PYR-1 | Mvan_1614 | - | 0.0 | 69.21% (945) | transport protein |
| M. vanbaalenii PYR-1 | Mvan_1936 | - | 0.0 | 70.05% (925) | transport protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0695c | mmpL5 | 0.0 | 75.66% (953) | transmembrane transport protein MmpL5 |
| M. gilvum PYR-GCK | Mflv_3435 | - | 0.0 | 93.28% (967) | transport protein |
| M. tuberculosis H37Rv | Rv0676c | mmpL5 | 0.0 | 75.55% (953) | transmembrane transport protein MmpL5 |
| M. leprae Br4923 | MLBr_02378 | - | 0.0 | 63.38% (942) | hypothetical protein MLBr_02378 |
| M. abscessus ATCC 19977 | MAB_4382c | - | 0.0 | 66.49% (967) | putative membrane protein, MmpL |
| M. marinum M | MMAR_1005 | mmpL5 | 0.0 | 74.87% (955) | transmembrane transport protein, MmpL5 |
| M. avium 104 | MAV_2510 | - | 0.0 | 71.38% (961) | MmpL4 protein |
| M. smegmatis MC2 155 | MSMEG_3496 | - | 0.0 | 74.59% (968) | MmpL4 protein |
| M. thermoresistible (build 8) | TH_3260 | - | 0.0 | 70.48% (962) | PUTATIVE MmpL4 protein |
| M. ulcerans Agy99 | MUL_0757 | mmpL5 | 0.0 | 74.66% (955) | transmembrane transport protein, MmpL5 |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_3189|M.vanbaalenii_PYR-1 MSAPVSDARTDEFPAA--RQPHRP------FIPRMIRLFAVPIILGWIAL
Mflv_3435|M.gilvum_PYR-GCK MSAPVSDARTDEFPAA--HGPHRS------FIPRMIRLFAVPIILGWIAL
Mb0695c|M.bovis_AF2122/97 MIVQRTAAPTGSVPPD--RHAARP------FIPRMIRTFAVPIILGWLVT
Rv0676c|M.tuberculosis_H37Rv MIVQRTAAPTGSVPPD--RHAARP------FIPRMIRTFAVPIILGWLVT
MMAR_1005|M.marinum_M MTATTTDTPTDVVPPA--EHPPRP------FIPRMIRTFAVPLILGWVLL
MUL_0757|M.ulcerans_Agy99 MTATTTDTPTDVVPPA--EHPPRP------FIPRMIRTFAVPLILGWVLL
TH_3260|M.thermoresistible__bu MTANPGHVRTDEAPTD--RLDIKVSTHGPSFISTWIRRLSLPIIVGWIAL
MAB_4382c|M.abscessus_ATCC_199 MSKPMNDPTTDAFPAA--RHAARP------WLPRMIRVLAVPIVLGWIAV
MAV_2510|M.avium_104 MSTTFDDTRTDAIPVV--KEPVRE------KIPRFIRAFAVPIIIGWIAL
MSMEG_3496|M.smegmatis_MC2_155 MSNPSHDAPTDAMGVSGPKHARPG------GIAKWIRVLAVPIILVWVAI
MLBr_02378|M.leprae_Br4923 MTVKCANDLDTHTKPP--------------FVARMIHRFAVPIILVWLAI
* :. *: :::*::: *:
Mvan_3189|M.vanbaalenii_PYR-1 IVILNVTVPQLEAVGEARAVSMSPNEAPSLISMKKVGELFREGDSDSSVM
Mflv_3435|M.gilvum_PYR-GCK IVVVNVTVPQLETVGEAQAVSMSPNDAPSMISMKKVGELFEEGDSDSSVM
Mb0695c|M.bovis_AF2122/97 IAVLNVTVPQLETVGQIQAVSMSPDAAPSMISMKHIGKVFEEGDSDSAAM
Rv0676c|M.tuberculosis_H37Rv IAVLNVTVPQLETVGQIQAVSMSPDAAPSMISMKHIGKVFEEGDSDSAAM
MMAR_1005|M.marinum_M IAVLNSVVPQLEVVGQMQAVSMSPDAAPSMISMKHIGKVFREGDSDSAAM
MUL_0757|M.ulcerans_Agy99 IAVLNSVVPQLEVVGQMQAVSMSPDAAPSMISMKHIGKVFREGDSDSAAM
TH_3260|M.thermoresistible__bu IALLNITVPQLEVVGEMRAVSMSPDEAPSMVAMKRVGELFDEGSSDSQVM
MAB_4382c|M.abscessus_ATCC_199 TIALNTVVPQLEEVGHMRAVSMSPNDAPAMIATKRVGEVFEEFKTSSSVM
MAV_2510|M.avium_104 IAVLNVVVPQLDEVGKMRSVEMTPDDAQSVVATKRMGAVFNEYKSNSSVM
MSMEG_3496|M.smegmatis_MC2_155 IAVLNTVVPQLDVVGEMRSVSMSPDDAPSVIAMKRVGEVFEEFKSNSSVM
MLBr_02378|M.leprae_Br4923 AVTVSVFIPSLEDVGQERSVSLSPKDAPSYIAMQKMGQVFNEGNSDSVIM
:. :*.*: **. ::*.::*. * : :: :::* :* * .:.* *
Mvan_3189|M.vanbaalenii_PYR-1 IVFEGDQPLGDEAHAWYDELVERLRADTKHVQSVQDFWSDPLTASGSQSN
Mflv_3435|M.gilvum_PYR-GCK IVFEGDQPLGDEAHAWYDELVDRLEADTAHVQSVQDFWSDPLTASGSQSN
Mb0695c|M.bovis_AF2122/97 IVLEGQRPLGDAAHAFYDQMIGRLQADTTHVQSLQDFWGDPLTATGAQSS
Rv0676c|M.tuberculosis_H37Rv IVLEGQRPLGDAAHAFYDQMIGRLQADTTHVQSLQDFWGDPLTATGAQSS
MMAR_1005|M.marinum_M IVLEGQEPLGAAAHKFYDKMITKLRADTKHVQSVQDFWGDPLTATGAQSG
MUL_0757|M.ulcerans_Agy99 IVLEGQEPLGAAAHKFYDKMITKLRADTKHVQSVQDFWGDPLTATGAQSG
TH_3260|M.thermoresistible__bu IVLEGEEPLGADARAFYDELVKRLEADDAHVENVQDLWSDPLTATGVQSP
MAB_4382c|M.abscessus_ATCC_199 IVLEGEQSLGADAHRFYDQMVRDLRADTTHVQHVQDFWADALTAAAAQSS
MAV_2510|M.avium_104 IVLEGQQPLGADAHAYYDEIVRRLNADTKHVEHVQDMWSDPLTGAGAQSN
MSMEG_3496|M.smegmatis_MC2_155 IVLEGEQPLGDEAHKYYDEIVDKLEADPAHVEHVQDFWGDPLTASGAQSP
MLBr_02378|M.leprae_Br4923 IVLEGDKPLGDDAHRFYDGLIRKLRAD-KHVQSVQDFWGDPLTAPGAQSN
**:**:..** *: :** :: *.** **: :**:*.*.**... **
Mvan_3189|M.vanbaalenii_PYR-1 DGKAAYVQVKLAGNQGESLANESVQAAQEIVRSLE---PPPGVRAFVTGP
Mflv_3435|M.gilvum_PYR-GCK DGKAAYVQVKLSGNQGEALANESVKAVQQTVSSLE---PPPGVRAFVTGP
Mb0695c|M.bovis_AF2122/97 DGKAAYVQVKLAGNQGESLANESVEAVKTIVERLA---PPPGVKVYVTGS
Rv0676c|M.tuberculosis_H37Rv DGKAAYVQVKLAGNQGESLANESVEAVKTIVERLA---PPPGVKVYVTGS
MMAR_1005|M.marinum_M DGKAAYVQVKLAGNQGESLANESVEAVKTIVDSLQ---APPGVKVYVTGS
MUL_0757|M.ulcerans_Agy99 DGKAAYVQVKLAGNQGESLANESVEAVKTIVDSLQ---APPGVKVYVTGS
TH_3260|M.thermoresistible__bu DGKSAYVQVKLAGNQGEALANESVETVREMIATVSEEMAPEGVQAFVTGG
MAB_4382c|M.abscessus_ATCC_199 DGKAAYVQVYIAGDQGEALANESVEAVRHITAAVP---PSPGVRVYVTGP
MAV_2510|M.avium_104 DGKASYVQVYLAGNQGEALANESVESVQNIVKSVQ---APNGVKAYVTGP
MSMEG_3496|M.smegmatis_MC2_155 DGLASYVQVYTRGNQGEALANESVEAVQDIVESVP---APPGVKAYVTGP
MLBr_02378|M.leprae_Br4923 DGKAAYVQLSLAGNQGELLAQESIDAVRKIVVQTP---APPGITAWVTGA
** ::***: *:*** **:**:.:.: .. *: .:***
Mvan_3189|M.vanbaalenii_PYR-1 AALAADQHIASDRSVRVIELVTFAVIIVMLLLVYRSIVTVLLTLVMVVLS
Mflv_3435|M.gilvum_PYR-GCK AALAADQHIAGDRSMRLIEAATFTVIIVMLLLVYRSIVTVLITLVMVVMS
Mb0695c|M.bovis_AF2122/97 AALVADQQQAGDRSLQVIEAVTFTVIIVMLLLVYRSIITSAIMLTMVVLG
Rv0676c|M.tuberculosis_H37Rv AALVADQQQAGDRSLQVIEAVTFTVIIVMLLLVYRSIITSAIMLTMVVLG
MMAR_1005|M.marinum_M AALVADQQLAGDRSLQLIEMVTFTVIIVMLLLVYRSIITAAIMLTMVVLG
MUL_0757|M.ulcerans_Agy99 AALVADQQLAGDRSLQLIEMVTFTVIIVMLLLVYRSIITAAIMLTMVVLG
TH_3260|M.thermoresistible__bu AAMSADQQEAGDSSLRVIEALTFLVITTMLLFVYRSVITVLIALFMVLIG
MAB_4382c|M.abscessus_ATCC_199 ATTSADQEIVANDSMELIELVTFGVIMVMLLLVYRSVITMVIVVAMVMLE
MAV_2510|M.avium_104 AALSADQHTAGDRSLQLITAATFTVIIGMLLLVYRSVITVLLTLVMVVLE
MSMEG_3496|M.smegmatis_MC2_155 AALSADQHVASDRSVRVIEALTFAVIITMLLLVYRSIVTVILTLVMVVLS
MLBr_02378|M.leprae_Br4923 SALIADMHHSGDKSMIRITATSVIVILVTLLLVYRSFITVILLLFTVGIE
:: ** . .: *: * :. ** **:****.:* : : * :
Mvan_3189|M.vanbaalenii_PYR-1 LATARGVVAFLGWHEIIGLSLFATNLLVTLAIAAATDYAIFLIGRYQEAR
Mflv_3435|M.gilvum_PYR-GCK LATARGVVAFLGHHEIIGLSTFATNLLVTLAIAAATDYAIFLIGRYQEAR
Mb0695c|M.bovis_AF2122/97 LLATRGGVAFLGFHRIIGLSTFATNLLVVLAIAAATDYAIFLIGRYQEAR
Rv0676c|M.tuberculosis_H37Rv LLATRGGVAFLGFHRIIGLSTFATNLLVVLAIAAATDYAIFLIGRYQEAR
MMAR_1005|M.marinum_M LLATRGGVAFLGYHRIIGLSTFATNLLVVLAIAAATDYAIFLIGRYQEAR
MUL_0757|M.ulcerans_Agy99 LLATRGGVAFLGYHRIIGLSTFATNLLVVLAIAAATDYAIFLIGRYQEAR
TH_3260|M.thermoresistible__bu LLTTRGVVAFLGYHNIIGLSTFATSLLVTLAIAIAVDYAIFLIGRYQEAR
MAB_4382c|M.abscessus_ATCC_199 LASARGLVALLSYHNAFGLTSFATSLVVTLAIAAATDYAIFLIGRYQEAR
MAV_2510|M.avium_104 LSAARGMVAFLGYYKIIGLSTFATNLLVTLAIAAATDYAIFLIGRYQEAR
MSMEG_3496|M.smegmatis_MC2_155 LSAARGMIAFLGYHEIIGLSVFATNLLTTLAIAAATDYAIFLIGRYQEAR
MLBr_02378|M.leprae_Br4923 SAVARGVVALLGHTGLIGLSTFAVNLLTSLAIAAGTDYGIFITGRYQEAR
:** :*:*. :**: **..*:. **** ..**.**: *******
Mvan_3189|M.vanbaalenii_PYR-1 TNGEDKESAYYTMFHGTAHVVLGSGLTIAGATYCLSFTRLPYFQTLGVPL
Mflv_3435|M.gilvum_PYR-GCK TAGEDKETAYYTMFHGTAHVVLGSGLTIAGATFCLSFTRLPYFQTMGVPL
Mb0695c|M.bovis_AF2122/97 GLGQDRESAYYTMFGGTAHVVLGSGLTIAGATFCLSFTRLPYFQTLGVPL
Rv0676c|M.tuberculosis_H37Rv GLGQDRESAYYTMFGGTAHVVLGSGLTIAGATFCLSFTRLPYFQTLGVPL
MMAR_1005|M.marinum_M GLGQDRESAYYTMFGGTAHVVLGSGLTIAGATFCLSFTRLPYFQTLGVPL
MUL_0757|M.ulcerans_Agy99 GLGQDRESAYYTMFGGTAHVVLGSGLTIAGATFCLSFTRLPYFQTLGVPL
TH_3260|M.thermoresistible__bu SVGADRATAYHTMFGGTAHVILASGLTIAGATFCLSFTRLPYFQSLGPPL
MAB_4382c|M.abscessus_ATCC_199 RAGEDRESAYYTMFHGTAHVVLASGLTIAGATFCLHFTRLPYFKTTGIPL
MAV_2510|M.avium_104 AVGESREDAYYTMYKGTAHVVAGSGMTIAGATFCLHFTNLPYFQTLGIPL
MSMEG_3496|M.smegmatis_MC2_155 SVGEDREQSYYTMFHSTAHVVLGSGMTIAGATLCLHFTRMPYFQSLGIPL
MLBr_02378|M.leprae_Br4923 QANENKEAAFYTMYRGTFHVILGSGLTISGATFCLSFARMPYFQTLGVPC
. .: :::**: .* **: .**:**:*** ** *:.:***:: * *
Mvan_3189|M.vanbaalenii_PYR-1 AIGMFVVVMAGVILMVAMISVATRFGKLLEPKRAMRIRGWRKIGAAVVRW
Mflv_3435|M.gilvum_PYR-GCK AIGMFVVVMAGVVLMVACISVATRFGKLLEPKRAMRIRGWRKIGAAIVRW
Mb0695c|M.bovis_AF2122/97 AIGMVIVVAAALTLGPAIIAVTSRFGKLLEPKRMARVRGWRKVGAAIVRW
Rv0676c|M.tuberculosis_H37Rv AIGMVIVVAAALTLGPAIIAVTSRFGKLLEPKRMARVRGWRKVGAAIVRW
MMAR_1005|M.marinum_M AIGMVIVVATALTLGPALIAVMSRFGKLLEPKRMARVRGWRKIGAAIVRW
MUL_0757|M.ulcerans_Agy99 AIGMVIVVATALTLGPALIAVMSRFGKLLEPKRMARVRGWRKIGAAIVRW
TH_3260|M.thermoresistible__bu AIGMIVLIVTSLTMGPAIITVATRFG-LLEPKRAARVRFWRKVGASVVRW
MAB_4382c|M.abscessus_ATCC_199 SIGMAIVVAMALTLGPALISVASRFGNVLEPKGAGKARGWRRVGAATVRW
MAV_2510|M.avium_104 AIGMVVVVAAALTLGPAVISVASRFRQTLEPRRTQRIRGWRKVGTVVVRW
MSMEG_3496|M.smegmatis_MC2_155 AIGMSVVVLASLTMGAAIISVASRFGKTFEPKRAMRTRGWRKLGAAVVRW
MLBr_02378|M.leprae_Br4923 AVGMLIAVAVALTLGPAVLTVGSRFG-LFEPKRLIKVRGWRRIGTVVVRW
::** : : .: : * ::* :** :**: : * **::*: ***
Mvan_3189|M.vanbaalenii_PYR-1 PGPILVATLAITLVGLLALPGYKTNYNDRTYLPADLPANEGYAVADRHFD
Mflv_3435|M.gilvum_PYR-GCK PGPILVATMAITLVGLLALPGYKTNYNDRDYLPADLPANQGYAVADEHFD
Mb0695c|M.bovis_AF2122/97 PGPILVGAVALALVGLLTLPGYRTNYNDRNYLPADLPANEGYAAAERHFS
Rv0676c|M.tuberculosis_H37Rv PGPILVGAVALALVGLLTLPGYRTNYNDRNYLPADLPANEGYAAAERHFS
MMAR_1005|M.marinum_M PGPILIGAVALALVGLLTLPGYKTNYNDRNYLPADLPANEGYSAAERHFS
MUL_0757|M.ulcerans_Agy99 PGPILIGAVALALVGLLTLPGYKTNYNDRNYLPADLPANEGYSAAERHFS
TH_3260|M.thermoresistible__bu PGPIMVTTVAVTLVGLLALPGYRTNYNDRNYLPASFPSNAGYEAAERHFD
MAB_4382c|M.abscessus_ATCC_199 PGAILVAAVTMALVGLLTLPGYHTTYNDRIYLPGQVPANVGYAAADRHFS
MAV_2510|M.avium_104 PGPILVLTIGVALVGLLTLPGYRTNYNDRNYLPTDLPANEGYAAADRHFS
MSMEG_3496|M.smegmatis_MC2_155 PAPILVTTIALSVVGLLALPGYQTNYNDRRYLPQDLPANTGYAAADRHFS
MLBr_02378|M.leprae_Br4923 PLPILITTCAIAMVGLLALPGYRTNYKDRAYLPASIPANQGFAAADRHFP
* .*:: : :::****:****:*.*:** *** ..*:* *: .*:.**
Mvan_3189|M.vanbaalenii_PYR-1 QARMNPELLMIESDHDLRNSADFLVIDKIAKAIFKVEGIARVQAITRPDG
Mflv_3435|M.gilvum_PYR-GCK QARMNPELLMIESDHDLRNSADFLVIDKIAKALFRVEGIARVQAITRPDG
Mb0695c|M.bovis_AF2122/97 QARMNPEVLMVESDHDMRNSADFLVINKIAKAIFAVEGISRVQAITRPDG
Rv0676c|M.tuberculosis_H37Rv QARMNPEVLMVESDHDMRNSADFLVINKIAKAIFAVEGISRVQAITRPDG
MMAR_1005|M.marinum_M QARMNPEVLMVESDHDMRNSSDFLVINKIAKAIFGVEGISRVQAITRPDG
MUL_0757|M.ulcerans_Agy99 QARMNPEVLMVESDHDMRNSSDFLVINKIAKAIFGVEGISRVQAITRPDG
TH_3260|M.thermoresistible__bu PARLNPELLLIESDHDLRNPADFLVIEKIAKAVFQVEGISRVQGITRPEG
MAB_4382c|M.abscessus_ATCC_199 KAKMNPDLLLVESDHDLRNPADFLVIDKIAKALARVRGIAQVQTITRPDG
MAV_2510|M.avium_104 QARMNPEVLMIESDHDLRNSADFLVIDKIAKTVFRVPGIGRVQAITRPQG
MSMEG_3496|M.smegmatis_MC2_155 QARMNPELLMIESDHDLRNSADFLVVDRIAKRVFQVPGISRVQAITRPQG
MLBr_02378|M.leprae_Br4923 QARMKPEILMIESDHDMRNPADFLILDKLARGIFRVPGISRVQAITRPDG
*:::*::*::*****:**.:***:::::*: : * **.:** ****:*
Mvan_3189|M.vanbaalenii_PYR-1 KPIKHTSIPFQMSMQGTTQRLNEKYMQDRMADMLVQADEMANTIATMEKM
Mflv_3435|M.gilvum_PYR-GCK KPIKHTSIPFQMSMQGTTQRLNEKYMQDRMADMLVQADEMANTIKTMEKM
Mb0695c|M.bovis_AF2122/97 KPIEHTSIPFLISMQGTSQKLTEKYNQDLTARMLEQVNDIQSNIDQMERM
Rv0676c|M.tuberculosis_H37Rv KPIEHTSIPFLISMQGTSQKLTEKYNQDLTARMLEQVNDIQSNIDQMERM
MMAR_1005|M.marinum_M KPIEHTSIPFLISMQGTSQKLTEKYNQDLTTSMLQQVNDIQTNIDQMERM
MUL_0757|M.ulcerans_Agy99 KPIEHTSIPFLISMQGTSQKLTEKYNQDLTTSMLQQVNDIQTNIDQMERM
TH_3260|M.thermoresistible__bu TPIEHSSIPFMISQQGTLQILNQQYMQDRMADMLVQADEMQLMIDTMDKM
MAB_4382c|M.abscessus_ATCC_199 KPIKHTSIPFNMSTANTAQTLTNDYSQANFANLLKQADAMQVTIDSMEKM
MAV_2510|M.avium_104 TPIEHTSIPFQISMQGVTQQMNQKYQQDQMADMLHQADMMQTTIDSMEKM
MSMEG_3496|M.smegmatis_MC2_155 TPIEHTSIPFQISMSGTTQMMNMKYMQDRMADMLVMADEMQKSVDTMEEM
MLBr_02378|M.leprae_Br4923 TAMDHTSIPFQISMQNAGQVQTMKYQKDRMNDLLRQAENMAETIASMRRM
..:.*:**** :* .. * . .* : :* .: : : * .*
Mvan_3189|M.vanbaalenii_PYR-1 SNLTAQMADITHSMVSKMENMLTDIEDLRDSIANFDDFFRPIRNYFYWEP
Mflv_3435|M.gilvum_PYR-GCK SGLTAQMADITHSMVTKMEGMVVDIEDLRDSIANFDDFFRPIRNYFYWEP
Mb0695c|M.bovis_AF2122/97 HSLTQQMADVTHEMVIQMTGMVVDVEELRNHIADFDDFFRPIRSYFYWEK
Rv0676c|M.tuberculosis_H37Rv HSLTQQMADVTHEMVIQMTGMVVDVEELRNHIADFDDFFRPIRSYFYWEK
MMAR_1005|M.marinum_M HDLTQQMADVTHQMVIQMKGMVVDVEELRNHIADFDDFFRPIRSYFYWEK
MUL_0757|M.ulcerans_Agy99 HDLTQQMADVTHQMVIQMKGMVVDVEELRNHIADFDDFFRPIRSYFYWEK
TH_3260|M.thermoresistible__bu IALMEEMVETTDNMVRDMDEMAAHVAEMRDSISEFDDFFRPIRNYFYWEP
MAB_4382c|M.abscessus_ATCC_199 QAITQQMAEVTASMSANMKNTAQDINQLRDNMSNFDDQFRPIRNYFYWEK
MAV_2510|M.avium_104 QSITVQMSNDMHVMVQKMHDMTIDINDLRNKMADFEDFFRPMRSYFYWEK
MSMEG_3496|M.smegmatis_MC2_155 LKITREMSDTTHSMVGKMHGMVEDIKELRDHIADFDDFLRPIRNYFYWEP
MLBr_02378|M.leprae_Br4923 HQLMALLTENTHHILNDTVEMQKTTSKLRDEIANFDDFWRPIRSYFYWER
: : : : . .:*: :::*:* **:*.*****
Mvan_3189|M.vanbaalenii_PYR-1 HCYNIPVCWALRSVFDTLDGINVMTDDFREILPDMKRLDSLMPQMVALMP
Mflv_3435|M.gilvum_PYR-GCK HCYNIPVCWALRSVFDTLDGINIMTDDFKAIIPDMKRLDTLMPQMVALMP
Mb0695c|M.bovis_AF2122/97 HCYDIPVCWSLRSVFDTLDGIDVMTEDINNLLPLMQRLDTLMPQLTAMMP
Rv0676c|M.tuberculosis_H37Rv HCYDIPVCWSLRSVFDTLDGIDVMTEDINNLLPLMQRLDTLMPQLTAMMP
MMAR_1005|M.marinum_M HCFDIPVCWSLRSVFDTLDGVDLMTEDINNLLPLMERLDTLMPQMTAMMP
MUL_0757|M.ulcerans_Agy99 HCFDIPVCWSLRSVFDTLDGVDLMTEDINNLLPLMERLDTLMPQMTAMMP
TH_3260|M.thermoresistible__bu HCYNIPVCWAMRSVFDTLDGINTMTDDIQNMLPHMHRLNDLMPQMLTLLP
MAB_4382c|M.abscessus_ATCC_199 HCFDIPMCWALRSVFDALDGISLMSDDFDTLVPDIQRMAGLMPQMVVLMP
MAV_2510|M.avium_104 HCYDIPVCWSLRSVFDALDGIDTMTDDIQSLLPIMDHLDTLMPQMVALMP
MSMEG_3496|M.smegmatis_MC2_155 HCDSIPVCQSIRSIFDTLDGIDVMTDDIQRLMPDMDRLDELMPQMLTIMP
MLBr_02378|M.leprae_Br4923 HCFNIPICWSFRSIFDALDGVDQIDERLNSIVGDIKNMDLLMPQMLEQFP
** .**:* ::**:**:***:. : : : :: :..: ****: :*
Mvan_3189|M.vanbaalenii_PYR-1 EMIETMKTMRTMMLTMYQSQKGQQDQMAAMSEDADAMGEAFDDSMNDDSF
Mflv_3435|M.gilvum_PYR-GCK EMIETMKTMRTMMLTMYQSQKGQQDQMAAMSEDADAMGEAFDDSMNDDSF
Mb0695c|M.bovis_AF2122/97 EMIQTMKSMKAQMLSMHSTQEGLQDQMAAMQEDSAAMGEAFDASRNDDSF
Rv0676c|M.tuberculosis_H37Rv EMIQTMKSMKAQMLSMHSTQEGLQDQMAAMQEDSAAMGEAFDASRNDDSF
MMAR_1005|M.marinum_M EMIQTMKSMKAQMLSMHSTQQGLQDQMAAMQEDSSAMGEAFDSSRNDDSF
MUL_0757|M.ulcerans_Agy99 EMIQTMKSMKAQMLSMHSTQQGLQDQMAAMQEDSSAMGEAFDSSRNDDSF
TH_3260|M.thermoresistible__bu TTVATMKTMRSMLLTMQATNAGLQDQMAEMQENANAMGEAFDQARNDDSF
MAB_4382c|M.abscessus_ATCC_199 PMIASMKEQKQIMLSQYQVQKAQQDQTMAMQANASAMGQAFDDAKNDDSF
MAV_2510|M.avium_104 SMIENMKAMKTTMLTMYSTQKGLQDQQNEAQKNSNAMGKAFDASKNDDSF
MSMEG_3496|M.smegmatis_MC2_155 PMIESMKTMKTMMLTTQATMGGLQDQMEAAMENQTAMGQAFDASKNDDSF
MLBr_02378|M.leprae_Br4923 PMIESMESMRTIMLTMHSTMSGIFDQMNELSDNANTMGKAFDTAKNDDSF
: .*: : :*: . ** : :**:*** : *****
Mvan_3189|M.vanbaalenii_PYR-1 YLPPEIFENKDFQRGLEQFLSPDGHAVRFIISHEGDPLSPEGVAKIDKIK
Mflv_3435|M.gilvum_PYR-GCK YLPPEIFENADFQRGLEQFLSPDGHAVRFIISHEGDPLSPEGVAKIDKIK
Mb0695c|M.bovis_AF2122/97 YLPPEVFDNPDFQRGLEQFLSPDGHAVRFIISHEGDPMSQAGIARIAKIK
Rv0676c|M.tuberculosis_H37Rv YLPPEVFDNPDFQRGLEQFLSPDGHAVRFIISHEGDPMSQAGIARIAKIK
MMAR_1005|M.marinum_M YLPPEVFNNPDFKRGLEQFLSPDGHAVRFIISHEGDPMSQAGINHIDKIK
MUL_0757|M.ulcerans_Agy99 YLPPEVFNNPDFKRGLEQFLSPDGHAVRFIISHEGDPMSQAGINHIDKIK
TH_3260|M.thermoresistible__bu YLPPEAFDNPDFQRGLEQFLSPDGHAVRFIISHQGDPMTPEGLAKIEPIK
MAB_4382c|M.abscessus_ATCC_199 YLPPEAFETADFKSGIKLFVSPNGHAVRFTIFHQSDPLTEEGTSRVEPLK
MAV_2510|M.avium_104 YLPPETFNNKEFKKGMKNFISPDGHAVRFIISHDGDPMTQEGISHIGAIK
MSMEG_3496|M.smegmatis_MC2_155 YLPPETFENPDFKRGMKMFLSPDGHAVRFIISHEGDPMSPEGIKHIDAIK
MLBr_02378|M.leprae_Br4923 YLPPEVFKNTDFKRAMKSFLSSDGHAARFIILHRGDPASVAGIASINAIR
***** *.. :*: .:: *:*.:***.** * * .** : * : ::
Mvan_3189|M.vanbaalenii_PYR-1 TAAKEAIKGTPLEGSKIYLGGTAATFKDMQDGNNYDLLIAGISALGLIFI
Mflv_3435|M.gilvum_PYR-GCK TAAKEAVKGTPLEGSKIYLGGTAAVFKDMQDGNNYDLLIAGIAALGLIFI
Mb0695c|M.bovis_AF2122/97 TAAKEAIKGTPLEGSAIYLGGTAAMFKDLSDGNTYDLMIAGISALCLIFI
Rv0676c|M.tuberculosis_H37Rv TAAKEAIKGTPLEGSAIYLGGTAAMFKDLSDGNTYDLMIAGISALCLIFI
MMAR_1005|M.marinum_M TAAKEAIKGTPLEGSTIYLGGTAAMFKDLSDGNTFDLMIAGISALCLIFI
MUL_0757|M.ulcerans_Agy99 TAAKEAIKGTPLEGSTIYLGGTVAMFKDLSDGNTFDLMIAGISALCLIFI
TH_3260|M.thermoresistible__bu KAAVEAIKGTPLEGSRVYLGGTAAVFKDMSDGTKYDLIIAAIAAVCLIFI
MAB_4382c|M.abscessus_ATCC_199 TAAEDAIKGTPLEGSSIYVGGSAALYKDMQEGADYDLLIAAVAALILIFL
MAV_2510|M.avium_104 KAAYEALKGTPLEGSKIYLAGTASIYKDLSDGNNYDLLIAGISSLCLIFI
MSMEG_3496|M.smegmatis_MC2_155 QAAKEAIKGTPLEGSKIYLGGTAATFKDLQEGANYDLIIAGIAALCLIFI
MLBr_02378|M.leprae_Br4923 TAAEEALKGTPLEDTKIYLAGTAAVFKDIDEGANWDLVIAGISSLCLIFI
** :*:******.: :*:.*:.: :**:.:* :**:**.:::: ***:
Mvan_3189|M.vanbaalenii_PYR-1 IMLILTRAIVAAAVIVGTVVLSLAASFGLSVLFWQHILGTELHWMVLAMA
Mflv_3435|M.gilvum_PYR-GCK IMLILTRAVVAAAVIVGTVVLSLAASFGLSVLFWQHILGQPLHWMVLAMA
Mb0695c|M.bovis_AF2122/97 IMLIITRSVVAAAVIVGTVVLSLGASFGLSVLIWQHILGIELHWLVLAMA
Rv0676c|M.tuberculosis_H37Rv IMLITTRSVVAAAVIVGTVVLSLGASFGLSVLIWQHILGIELHWLVLAMA
MMAR_1005|M.marinum_M IMLIITRSVVAAAVIVGTVVLSLGASFGLSVLIWQHILGIELHWLVLAMA
MUL_0757|M.ulcerans_Agy99 IMLIITRSVVAAAVIVGTVVLSLGASFGLSVLIWQHVLGIELHWLVLAMA
TH_3260|M.thermoresistible__bu IMLILTRAVVAAAVIVGTVTLSLAASFGVSVLIWQYIVGLEMHWMVFPMA
MAB_4382c|M.abscessus_ATCC_199 IMVLLTRAVVAAAVIVGTVVLSLGASFGLSVLLWQHIIGTPLHWMVLPMA
MAV_2510|M.avium_104 IMLIITRGVVASAVIVGTVLLSLGASFGLSVLIWQHLIGIELHWMVLAMS
MSMEG_3496|M.smegmatis_MC2_155 IMLIITRAVVASAVIVGTVVISLGASFGLSVLIWQHIIGLELHWMVLAMA
MLBr_02378|M.leprae_Br4923 IMLIITRAFVAAAVIVGTVALSLGASFGLSVLLWQHILGIELHYLVLAMS
**:: **..**:******* :**.****:***:**:::* :*::*:.*:
Mvan_3189|M.vanbaalenii_PYR-1 VIVLLAVGADYNLLLVSRLKEEIHAGIGTGIIRAMGGSGSVVTAAGLVFA
Mflv_3435|M.gilvum_PYR-GCK VIILLAVGADYNLLLVSRLKEEIHAGIGTGIIRAMGGSGSVVTAAGLVFA
Mb0695c|M.bovis_AF2122/97 VIILLAVGADYNLLLVARLKEEIHAGINTGIIRAMGGSGSVVTAAGLVFA
Rv0676c|M.tuberculosis_H37Rv VIILLAVGADYNLLLVARLKEEIHAGINTGIIRAMGGSGSVVTAAGLVFA
MMAR_1005|M.marinum_M VIILLAVGADYNLLLVARLKEEIHAGINTGIIRAMGGSGSVVTAAGLVFA
MUL_0757|M.ulcerans_Agy99 VIILLAVGADYNLLLVARLKEEIHAGINTGIIRAMGGSGSVVTAAGLVFA
TH_3260|M.thermoresistible__bu LIILLAVGADYNLLVVARLREEIPAGLNTGMIRAMGGSGSVVTAAGLVFG
MAB_4382c|M.abscessus_ATCC_199 VIILLAVGADYNLLLVSRMKEELHAGLHTGTIRSMAGTGSVVTSAGLVFA
MAV_2510|M.avium_104 VIILLAVGADYNLLLVARFKEEIHAGLNTGIIRSMGGTGSVVTSAGLVFA
MSMEG_3496|M.smegmatis_MC2_155 VIVLLAVGADYNLLLVSRIKEEIHAGLNTGIIRSMGGTGSVVTSAGLVFA
MLBr_02378|M.leprae_Br4923 VIVLLAVGSDYNLLLVSRFKQEIQAGLKTGIIRSMGGTGKVVTNAGLVFA
:*:*****:*****:*:*:::*: **: ** **:*.*:*.*** *****.
Mvan_3189|M.vanbaalenii_PYR-1 LTMMSMAVSELTVIGQVGTTIGLGLLFDTLVIRAFMTPSIAALLGPWFWW
Mflv_3435|M.gilvum_PYR-GCK LTMMSMSVSELTVIGQVGTTIGLGLLFDTLVIRAFMTPSIAALLGPWFWW
Mb0695c|M.bovis_AF2122/97 FTMMSFAVSELTVMAQVGTTIGMGLLFDTLIVRSFMTPSIAALLGKWFWW
Rv0676c|M.tuberculosis_H37Rv FTMMSFAVSELTVMAQVGTTIGMGLLFDTLIVRSFMTPSIAALLGKWFWW
MMAR_1005|M.marinum_M FTMMSFAVSELTVMAQVGTTIGMGLLFDTLIVRSFMTPSIAALLGKWFWW
MUL_0757|M.ulcerans_Agy99 FTMMSFAVSELTVMAQVGTTIGMGLLFDTLIVRSFMTPSIAALLGKWFWW
TH_3260|M.thermoresistible__bu LTMMVMVVSDLTVMGQVGTTIGLGLLFDTFVVRSFMMPSIATLLGRWFWW
MAB_4382c|M.abscessus_ATCC_199 FTMMGMMVSSFTVIGQIGTTIGLGLLFDTLVVRSFMTPSIASLLGRWFWW
MAV_2510|M.avium_104 FTMMTMAVSELTVIGQVGTTIGLGLLFDTLIVRSLMTPSIAALLGKWFWW
MSMEG_3496|M.smegmatis_MC2_155 FTMMSMAVSELAVIAQVGTTIGLGLLFDTLVIRSFMTPSIAALMGKWFWW
MLBr_02378|M.leprae_Br4923 FTMASMVVSDLRVIGQVGTTIGLGLLFDTLIVRSFMMPSIAALLGRWFWW
:** : **.: *:.*:*****:******:::*::* ****:*:* ****
Mvan_3189|M.vanbaalenii_PYR-1 PQRVRTRPVPAPWPRPGGLQSDPSEGVKV
Mflv_3435|M.gilvum_PYR-GCK PQRVRTRPVPAPWPRSGGLQSDPSEGVRA
Mb0695c|M.bovis_AF2122/97 PQVVRQRPVPQPWPSPASARTFALV----
Rv0676c|M.tuberculosis_H37Rv PQVVRQRPIPQPWPSPASARTFALV----
MMAR_1005|M.marinum_M PQVVRQRPAPQPWPTPAPTP--ATTSV--
MUL_0757|M.ulcerans_Agy99 PQVVRQRPAPQPWPTPAPTPPPATTSV--
TH_3260|M.thermoresistible__bu PQFVRKRPVPQPWPSVPPRGPATTDRVSV
MAB_4382c|M.abscessus_ATCC_199 PMVVRPRPRPQPWPQPLHAGAAEAVRA--
MAV_2510|M.avium_104 PQRVRQRPVPSPWPKPAAQEREVLADA--
MSMEG_3496|M.smegmatis_MC2_155 PQRVRQRPLPAPWPQPVQRDPEDALNRV-
MLBr_02378|M.leprae_Br4923 PQQGRTRPLLTVAAPVGLPPDRSLVYSD-
* * ** .