For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LRAVVLRDDRLQVRETPDPEPGPGELLLRTLSTAICASDVHFMDHPELALDDPTGRSLYDSGRDIVLGHE FVGEVVGHGPGCTGQFAIGTRVTAMPVRLVDGGAGGMRIIGQHPDAQGSFAELLVVSEAAAKPVPGEVSS DAVAVTDAFAVGEFYVRSARLQPGEVAIVIGAGAIGLSAVAALAARGVEPIIVADFRADRRRLAADSFGA HLLVDPREESPFAAFNAARARYGIPGPAAVFECVGAPGLIQNLVESAEMGTRIYCAGGWYTGDTLDITTA TRQGVTIQFGGGPHPQDWYGTLDAIVAGRLDPLPSVGKIIGLDEVPDALELARRSDGPPRIIVHPNGDTR
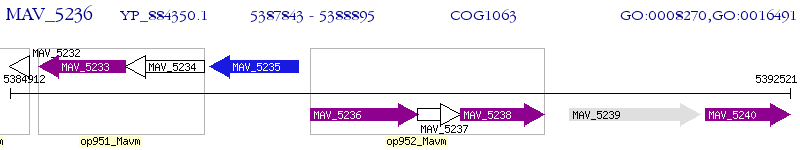
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_5236 | - | - | 100% (350) | oxidoreductase, zinc-binding dehydrogenase family protein |
| M. avium 104 | MAV_0537 | - | 5e-46 | 34.38% (352) | zinc-binding dehydrogenase |
| M. avium 104 | MAV_4173 | - | 2e-20 | 26.65% (349) | oxidoreductase, zinc-binding dehydrogenase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3753 | - | 6e-45 | 31.71% (391) | putative dehydrogenase |
| M. gilvum PYR-GCK | Mflv_3479 | - | 3e-19 | 26.39% (341) | alcohol dehydrogenase |
| M. tuberculosis H37Rv | Rv3726 | - | 6e-45 | 31.71% (391) | dehydrogenase |
| M. leprae Br4923 | MLBr_00118 | - | 7e-08 | 23.14% (242) | putative oxidireductase |
| M. abscessus ATCC 19977 | MAB_0526c | - | 6e-44 | 33.33% (336) | hypothetical protein MAB_0526c |
| M. marinum M | MMAR_3328 | adhE2_1 | 2e-51 | 35.15% (367) | zinc-dependent alcohol dehydrogenase AdhE2_1 |
| M. smegmatis MC2 155 | MSMEG_6108 | - | 1e-51 | 35.41% (353) | zinc-binding dehydrogenase |
| M. thermoresistible (build 8) | TH_4631 | - | 3e-12 | 26.75% (228) | PUTATIVE Alcohol dehydrogenase GroES domain protein |
| M. ulcerans Agy99 | MUL_2962 | - | 8e-16 | 25.50% (349) | dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_5762 | - | 1e-39 | 29.61% (385) | alcohol dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3753|M.bovis_AF2122/97 MKAVTCTNAKLEVVD-RPSPA-PAKGQLLLDVLRCGICGSDLHARLHCDE
Rv3726|M.tuberculosis_H37Rv MKAVTCTNAKLEVVD-RPSPA-PAKGQLLLDVLRCGICGSDLHARLHCDE
Mvan_5762|M.vanbaalenii_PYR-1 MKAVSCVHGTLEVVD-LPSPR-PGEGQLVLDVLRCGICGSDLHAKDHADE
MMAR_3328|M.marinum_M MRAVLMQDSQLQVAE-VPEPV-PGPGEVLVDVLACGICGSDLHCAAHGPE
MAB_0526c|M.abscessus_ATCC_199 MRASVLRNGSMVYRDDVPSPT-PGEGQVLVKVSACGICGSDLHFAKHGAR
MSMEG_6108|M.smegmatis_MC2_155 MRAAVLRQGRMVVRDDVAEPV-PGPGQVLVEVKACGICGSDLHFAQHGED
MAV_5236|M.avium_104 MRAVVLRDDRLQVRE-TPDPE-PGPGELLLRTLSTAICASDVHFMDHPEL
Mflv_3479|M.gilvum_PYR-GCK MKSTIVTGPGSTEVIDITRPQ-CGPDDVLVRMRACGICGSDAFYITIG--
MUL_2962|M.ulcerans_Agy99 MRTVVIDGPGSIRVDNRPDPALPGSDGVIVVVSAAGICGSDLHFYEGEYP
TH_4631|M.thermoresistible__bu VNGXXXEETRRGRYNLCPHMR-----------------------------
MLBr_00118|M.leprae_Br4923 MHAIVAESANQMVWREVPDVT-AGPGEVLIEVAASGVNRADVLQVAG---
:. .
Mb3753|M.bovis_AF2122/97 LADVMAESG----YHAFMRSNQQVVFGHEFCGEVVDYGPGTRRTPRRGTP
Rv3726|M.tuberculosis_H37Rv LADVMAESG----YHAFMRSNQQVVFGHEFCGEVVDYGPGTRRTPRRGTP
Mvan_5762|M.vanbaalenii_PYR-1 LTEVMAEGG----YHDFVRSDTPVVMGHEFCGVVADRGRKVGKEFKPGST
MMAR_3328|M.marinum_M FNSATQAAG----GFDMMDLSRPVVFGHEFVGRIAAYGPDTVQRIPVGRR
MAB_0526c|M.abscessus_ATCC_199 AIELSKQMVGMPSLTDGVDLSTDVFMGHEFAAEVIEAGPRT-QAPASGTA
MSMEG_6108|M.smegmatis_MC2_155 LLASTAQLEGVANQNDDVDLSRDIFMGHEFSAEILEAGPST-DTYAPGTL
MAV_5236|M.avium_104 ALDDPTGRS-------LYDSGRDIVLGHEFVGEVVGHGPGCTGQFAIGTR
Mflv_3479|M.gilvum_PYR-GCK --GIPPRQG-------------HTPLGHEPAGEVVEVGAAVTGVDVGDHV
MUL_2962|M.ulcerans_Agy99 LAEPVALGH----------EAVGTVVEKGPDVHTVEVGDQVMVSSVAGCG
TH_4631|M.thermoresistible__bu --------------------------------------------------
MLBr_00118|M.leprae_Br4923 --KYPHPPG------------ASAIIGMEVAGVVAAVSSGVTEWSVG---
Mb3753|M.bovis_AF2122/97 VVAMPLLRRGNKE--VHGIGLSTMAPGAYAERLVVEQS----LTFPVPNG
Rv3726|M.tuberculosis_H37Rv VVAMPLLRRGNKE--VHGIGLSTMAPGAYAERLVVEQS----LTFPVPNG
Mvan_5762|M.vanbaalenii_PYR-1 VVAFPLMR-AHGG--VHLTGLSPLAPGGYAEQVVAEAA----LTFVVPNG
MMAR_3328|M.marinum_M VVSAPFLLREQKV--TLGFAG-PEAPGGYSERMLLSEP----LLIEVPDH
MAB_0526c|M.abscessus_ATCC_199 VTSIPILLSAKG---LEPIVYSNTTLGGYAEQMLLSAP----LLLPIPNG
MSMEG_6108|M.smegmatis_MC2_155 VTSVPVLISPQG---MDMIVYSNTTLGGYAERMLLSAP----LLLPVPNG
MAV_5236|M.avium_104 VTAMPVRLVDGGAGGMRIIGQHPDAQGSFAELLVVSEA----AAKPVPGE
Mflv_3479|M.gilvum_PYR-GCK VINPMAAPSG--------IIGNGGATGALADYLLIENAQRGKSLEVIPKH
MUL_2962|M.ulcerans_Agy99 ACAGCATRDPAMCVSGPQIFGSGALGGAQADLLAVPAT--DFQVLKIPAA
TH_4631|M.thermoresistible__bu ------------------FYGTPPVDGALCEYVTIGAA----FAHPVPAE
MLBr_00118|M.leprae_Br4923 -----------------QEVCALLAGGGYAEYVAVPAS----QVMPIPKG
*. .: : . :*
Mb3753|M.bovis_AF2122/97 LAPEIAALTEP-MAVGWHAVRRGEVGKGDVAIVIGCGPIGLAVICMLKSR
Rv3726|M.tuberculosis_H37Rv LAPEIAALTEP-MAVGWHAVRRGEVGKGDVAIVIGCGPIGLAVICMLKSR
Mvan_5762|M.vanbaalenii_PYR-1 LDPATAALTEP-MAVAYHAVRRSDVTTKDVAIVLGCGPVGLAVICHLKAL
MMAR_3328|M.marinum_M VPTEVAALTEP-LAVAYRTVARVDTTTDDVALVVGCGPIGLAVIAVLKMK
MAB_0526c|M.abscessus_ATCC_199 LDPRHAALTEP-MAVGLHAVNKSAIQPGTGAIVIGCGPVGIAVIAALHNL
MSMEG_6108|M.smegmatis_MC2_155 LDAGPAALTEP-MAVGLHAVNTSRIERNETALVIGCGPVGIAVIAALKLL
MAV_5236|M.avium_104 VSSDAVAVTDA-FAVGEFYVRSARLQPGEVAIVIGAGAIGLSAVAALAAR
Mflv_3479|M.gilvum_PYR-GCK IPWEVAALNEP-MAVARHGVNRCRPRPEHQVVVFGAGPIGLGATLAFKAL
MUL_2962|M.ulcerans_Agy99 ITTEQALLLTDNLATGWAAARRADIPFGATVAVIGLGAVGLCAVRSALAQ
TH_4631|M.thermoresistible__bu MTDNAAALCEP-LSVAIATVDKAAVAGGSRVLIAGAGPIGLMTAQVARAY
MLBr_00118|M.leprae_Br4923 VNVVDSAGIPEVACTVWSNLVMAAHLSAGQLLLLHGGTSGIGSHGIQVAR
: .. : *. *:
Mb3753|M.bovis_AF2122/97 GVHTVIASDFSPGRRALATA-CGADSVVDPVQDSPYAVAAGLGQGNRHLQ
Rv3726|M.tuberculosis_H37Rv GVHTVIASDFSPGRRALATA-CGADSVVDPVQDSPYAVAAGLGQGNRHLQ
Mvan_5762|M.vanbaalenii_PYR-1 GVRTIVASDFSPGRRALAAR-CGADVVVDPAVDSPYEAVPAKQRG---LT
MMAR_3328|M.marinum_M GVGPIVASDFSPERRALATQ-LGADVVLDPNENSPFEAF-----------
MAB_0526c|M.abscessus_ATCC_199 GIEPIVASDYSSARRDLAQR-MGAHQVVDPKIEPTFHAWSRIGAG-----
MSMEG_6108|M.smegmatis_MC2_155 GVEDIVASDFSPTRRRLAAA-MGAHVTLDPAQDSPFHQCR----------
MAV_5236|M.avium_104 GVEPIIVADFRADRRRLAADSFGAHLLVDPREESPFAAFNAARAR-----
Mflv_3479|M.gilvum_PYR-GCK GVRHVVVVDIVASRLAKALQ-IGADAVIDSSEEDVVARLIELHGE-----
MUL_2962|M.ulcerans_Agy99 GAATVFAVDRVQGRLQRAAE-WGATAVASPAAEAIVAATSG---------
TH_4631|M.thermoresistible__bu GATDIVVTDLDPHRRRLAHR-FGATTTLDPQTDDVTGLR-----------
MLBr_00118|M.leprae_Br4923 ALGAKVAVTAGSEEKLEFCRALGAQITINYRDEDFVARLQQQNGG-----
. .. . ** . :
Mb3753|M.bovis_AF2122/97 SILDAFDLAVGTVESLQRLRLPWWHLWRAAEAAGAATPKRP-VIFECVGV
Rv3726|M.tuberculosis_H37Rv SILDAFDLAVGTVERLQRLRLPWWHLWRAAEAAGAATPKRP-VIFECVGV
Mvan_5762|M.vanbaalenii_PYR-1 ELPALYELGVGSMEKLRKIPG-WSHVYRVADRLGGTAPKRP-VIFECVGV
MMAR_3328|M.marinum_M -------MAAAVTDDPARMAP--------AHTVFGQLPLRPTVAFECVGV
MAB_0526c|M.abscessus_ATCC_199 ----------------------------------------APVIFEAVGV
MSMEG_6108|M.smegmatis_MC2_155 ----------------------------------------PAVVFEAVGA
MAV_5236|M.avium_104 -----------------------------------YGIPGPAAVFECVGA
Mflv_3479|M.gilvum_PYR-GCK ------------------------------GEAMWPGKAGTDIYLDAAGA
MUL_2962|M.ulcerans_Agy99 --------------------------------------RGADSVIDAVDT
TH_4631|M.thermoresistible__bu ----------------------------------------VDAFIDASGA
MLBr_00118|M.leprae_Br4923 ----------------------------------------ADVILDIMGA
:: ..
Mb3753|M.bovis_AF2122/97 PGIIDGIIASAPLFSRVVVVGVC-MGSDHIRPAMAINK-----EINLRFV
Rv3726|M.tuberculosis_H37Rv PGIIDGIIASAPLFSRVVVVGVC-MGSDHIRPAMAINK-----EINLRFV
Mvan_5762|M.vanbaalenii_PYR-1 PGMIDGIIGAAPLASRVVVVGVC-MGDDKLRPAMAIGK-----EIDLRFV
MMAR_3328|M.marinum_M PGLIQQLIAGVPPGTRIGVAGIN-MSSDHFEPAQCIAK-----ELDIRFS
MAB_0526c|M.abscessus_ATCC_199 PGILNEVLRDAPRSARVVVVGVC-MEPDAITPYFGIAK-----EVAVQFV
MSMEG_6108|M.smegmatis_MC2_155 PGIIDDILRRCPAGTRVVVAGVC-MQPDTVHPLYAAVK-----QISVSFV
MAV_5236|M.avium_104 PGLIQNLVESAEMGTRIYCAGGW-YTGDTLDITTATRQ-----GVTIQFG
Mflv_3479|M.gilvum_PYR-GCK GAVVTTALAAAKAGAILGVVGVH-KEPVPVDFAGIMSN-----EITIVGS
MUL_2962|M.ulcerans_Agy99 DASMTDALNAVRAGGTVSVVGVHDLQPFPLPALTCLLR-----SITLRLT
TH_4631|M.thermoresistible__bu PAAVMSGLAAVRPAGRAVLVGMG-AETMELPVQTIQNR-----ELILTGV
MLBr_00118|M.leprae_Br4923 -AYLDRNIDALAIDGQLIVIGLQGGVKGELNLGKVLSKRARIIGSTLRAR
. : : * . . :
Mb3753|M.bovis_AF2122/97 LGYTPLEFRDTLHMLAD------GKVNAAPLITGTVGLPGVAAAFDALGD
Rv3726|M.tuberculosis_H37Rv LGYTPLEFRDTLHMLAD------GKVNAAPLITGTVGLPGVAAAFDALGD
Mvan_5762|M.vanbaalenii_PYR-1 FGYTPLEFRDTLHMLAD------GKLDASALVTGTVGLGGVAAAFDALGD
MMAR_3328|M.marinum_M LYYTLEEFAQTFAHLAA------GDFDVAPLLTGTVALEGVADAFERLRN
MAB_0526c|M.abscessus_ATCC_199 LAYEPAEFSETLRRIAH------GEIDVAPLVTGEVGLEDVGSAFADLGD
MSMEG_6108|M.smegmatis_MC2_155 FGYDPAEFASSLRAIAE------GDIDVRPMITGSVPLDGMADAFDELAT
MAV_5236|M.avium_104 GGPHPQDWYGTLDAIVA------GRLDPLPSVGKIIGLDEVPDALELARR
Mflv_3479|M.gilvum_PYR-GCK MG-YPEEIFQVTEDLVE------NWEKYALIVSHTIPFDKVHDALELADT
MUL_2962|M.ulcerans_Agy99 IAPVQRTWSELIPLLES------GRLDVDGIFTTSLPLDEAAKGYSIAGS
TH_4631|M.thermoresistible__bu FR-YANTWPAAIALIRT------GRVDVDALITGRYPLEKTAEALESDRI
MLBr_00118|M.leprae_Br4923 PVNGPNSKSEIVAAVTASIWPMIADGSVRPIIGARMAVQQAADAHQLLLS
: . . . .
Mb3753|M.bovis_AF2122/97 -PEAHAKIMIDPKSNAASPQPFRVE
Rv3726|M.tuberculosis_H37Rv -PEAHAKIMIDPKSNAASPQPFRVE
Mvan_5762|M.vanbaalenii_PYR-1 -PETHAKILIDPAGTAVTP------
MMAR_3328|M.marinum_M -NPHDAKILLDPTR-----------
MAB_0526c|M.abscessus_ATCC_199 -PERHCKILVVP-------------
MSMEG_6108|M.smegmatis_MC2_155 -PDAHCKILVTP-------------
MAV_5236|M.avium_104 -SDGPPRIIVHPNGDTR--------
Mflv_3479|M.gilvum_PYR-GCK -PGGAGKVVVTFD------------
MUL_2962|M.ulcerans_Agy99 RSGNDVKVLLKV-------------
TH_4631|M.thermoresistible__bu PGNIKSVVEVGTS------------
MLBr_00118|M.leprae_Br4923 -GKVSGKIVLTVAGSGTETLLSRHH
: :