For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRAVLMQDSQLQVAEVPEPVPGPGEVLVDVLACGICGSDLHCAAHGPEFNSATQAAGGFDMMDLSRPVVF GHEFVGRIAAYGPDTVQRIPVGRRVVSAPFLLREQKVTLGFAGPEAPGGYSERMLLSEPLLIEVPDHVPT EVAALTEPLAVAYRTVARVDTTTDDVALVVGCGPIGLAVIAVLKMKGVGPIVASDFSPERRALATQLGAD VVLDPNENSPFEAFMAAAVTDDPARMAPAHTVFGQLPLRPTVAFECVGVPGLIQQLIAGVPPGTRIGVAG INMSSDHFEPAQCIAKELDIRFSLYYTLEEFAQTFAHLAAGDFDVAPLLTGTVALEGVADAFERLRNNPH DAKILLDPTR
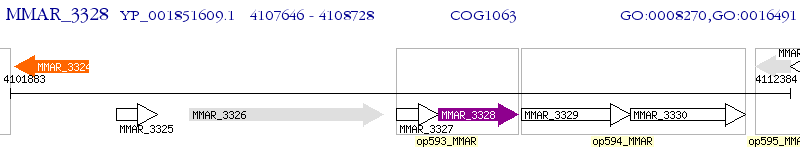
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3328 | adhE2_1 | - | 100% (360) | zinc-dependent alcohol dehydrogenase AdhE2_1 |
| M. marinum M | MMAR_5116 | - | 5e-75 | 44.75% (362) | dehydrogenase |
| M. marinum M | MMAR_3529 | - | 2e-26 | 27.99% (343) | dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3753 | - | 8e-79 | 43.12% (385) | putative dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0660 | - | 2e-28 | 28.53% (347) | alcohol dehydrogenase |
| M. tuberculosis H37Rv | Rv3726 | - | 1e-78 | 43.38% (385) | dehydrogenase |
| M. leprae Br4923 | MLBr_01784 | adhE2 | 1e-11 | 25.97% (231) | putative alcohol dehydrogenase (Zn dependent) |
| M. abscessus ATCC 19977 | MAB_0526c | - | 2e-62 | 39.94% (363) | hypothetical protein MAB_0526c |
| M. avium 104 | MAV_0537 | - | 4e-76 | 44.75% (362) | zinc-binding dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6108 | - | 3e-74 | 43.25% (363) | zinc-binding dehydrogenase |
| M. thermoresistible (build 8) | TH_4349 | - | 7e-17 | 29.72% (249) | PUTATIVE Alcohol dehydrogenase GroES domain protein |
| M. ulcerans Agy99 | MUL_4777 | - | 2e-17 | 27.30% (370) | zinc-dependent alcohol dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_5762 | - | 1e-78 | 45.00% (380) | alcohol dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3753|M.bovis_AF2122/97 ----MKAVTCTNAK-LEVVD-RPSPAPAKGQLLLDVLRCGICGSDLHARL
Rv3726|M.tuberculosis_H37Rv ----MKAVTCTNAK-LEVVD-RPSPAPAKGQLLLDVLRCGICGSDLHARL
Mvan_5762|M.vanbaalenii_PYR-1 ----MKAVSCVHGT-LEVVD-LPSPRPGEGQLVLDVLRCGICGSDLHAKD
MAV_0537|M.avium_104 ----MRAAVLRDGR-MVYRDDVPEPIPETGQVLVGVRACGICGSDLHFAA
MSMEG_6108|M.smegmatis_MC2_155 ----MRAAVLRQGR-MVVRDDVAEPVPGPGQVLVEVKACGICGSDLHFAQ
MAB_0526c|M.abscessus_ATCC_199 ----MRASVLRNGS-MVYRDDVPSPTPGEGQVLVKVSACGICGSDLHFAK
MMAR_3328|M.marinum_M ----MRAVLMQDSQ-LQVAE-VPEPVPGPGEVLVDVLACGICGSDLHCAA
Mflv_0660|M.gilvum_PYR-GCK ----MEAIVLNGTN-DVGLTSVPDPAPQDGEVIIEVAATGLCGTDLHEYV
TH_4349|M.thermoresistible__bu ----MKALRLMDWKSEPELVEVPDPTPGPGQVLVKIGGAGACHSDLHLMH
MUL_4777|M.ulcerans_Agy99 MAATMRAERFYADSKTVVLEDVPIPEPGPGEVLVKVAYCGICHSDLSLIN
MLBr_01784|M.leprae_Br4923 MSQTVRGVISRKKDEPVELVDIVIPDPGPGEAVVDVTACGVCHTDLTYRE
:.. * * *: :: : * * :**
Mb3753|M.bovis_AF2122/97 HCDELADVMAESG----YHAFMRSNQQVVFGHEFCGEVVDYGPGTRRTPR
Rv3726|M.tuberculosis_H37Rv HCDELADVMAESG----YHAFMRSNQQVVFGHEFCGEVVDYGPGTRRTPR
Mvan_5762|M.vanbaalenii_PYR-1 HADELTEVMAEGG----YHDFVRSDTPVVMGHEFCGVVADRGRKVGKEFK
MAV_0537|M.avium_104 HGAQVLQMSDRVAGGAAGMR-VDLDHDIFMGHEFSAEILEAGPDT-ETHP
MSMEG_6108|M.smegmatis_MC2_155 HGEDLLASTAQLEGVANQNDDVDLSRDIFMGHEFSAEILEAGPST-DTYA
MAB_0526c|M.abscessus_ATCC_199 HGARAIELSKQMVGMPSLTDGVDLSTDVFMGHEFAAEVIEAGPRT-QAPA
MMAR_3328|M.marinum_M HGPEFNSATQAAG----GFDMMDLSRPVVFGHEFVGRIAAYGPDTVQRIP
Mflv_0660|M.gilvum_PYR-GCK AGPTFSQPPVVLG-------HEVSGRIVEVGAGVDQSRIGEGAAVIPMDF
TH_4349|M.thermoresistible__bu DFEPGMMPWQLPFT----LGHENAGWVAAVGAGVGTVTEGDAVAVYGPWG
MUL_4777|M.ulcerans_Agy99 GTFPTQAPVVTQG-------HEASGTIVKLGPEVDGWSEGDRVVVAAGRP
MLBr_01784|M.leprae_Br4923 GGINDRYPFLLGH--------EAAGTVEVVGPGVTAVEPGDFVILNWRAV
. .* .
Mb3753|M.bovis_AF2122/97 RGTPVVAMP-----LLRRGNKEVHGIGLS------TMAPGAYAERLVVEQ
Rv3726|M.tuberculosis_H37Rv RGTPVVAMP-----LLRRGNKEVHGIGLS------TMAPGAYAERLVVEQ
Mvan_5762|M.vanbaalenii_PYR-1 PGSTVVAFP-----LMR-AHGGVHLTGLS------PLAPGGYAEQVVAEA
MAV_0537|M.avium_104 PGTLVTSLP-----VLL-SAKGVEPIVYS------NTTIGGYAERMLLSA
MSMEG_6108|M.smegmatis_MC2_155 PGTLVTSVP-----VLI-SPQGMDMIVYS------NTTLGGYAERMLLSA
MAB_0526c|M.abscessus_ATCC_199 SGTAVTSIP-----ILL-SAKGLEPIVYS------NTTLGGYAEQMLLSA
MMAR_3328|M.marinum_M VGRRVVSAP-----FLL-REQKVTLGFAG------PEAPGGYSERMLLSE
Mflv_0660|M.gilvum_PYR-GCK CGSCHYCHR-----SLY--HLCQRPGWIG------FTRNGGLANYVAVPS
TH_4349|M.thermoresistible__bu CGTCERCMGGAENYCENPAGAPVLSGGAG------LGMDGGMAEYLLVPA
MUL_4777|M.ulcerans_Agy99 CQSCPNCRR-------GDTVNCLRIQLMA------FAYDGAWAEYTVAQA
MLBr_01784|M.leprae_Br4923 CGQCRACKRGRPSYCFDTFNAQQKMTLIDGTELTPALGIGAFADKTLVHS
. *. ::
Mb3753|M.bovis_AF2122/97 S-LTFPVPNGLA-PEIAALTEPMAVGWHAVRR--GEVGKGDVAIVIGCGP
Rv3726|M.tuberculosis_H37Rv S-LTFPVPNGLA-PEIAALTEPMAVGWHAVRR--GEVGKGDVAIVIGCGP
Mvan_5762|M.vanbaalenii_PYR-1 A-LTFVVPNGLD-PATAALTEPMAVAYHAVRR--SDVTTKDVAIVLGCGP
MAV_0537|M.avium_104 P-LLLPIPNGLD-VKHAALTEPMAVGLHAVNK--SNITPAETALVLGCGP
MSMEG_6108|M.smegmatis_MC2_155 P-LLLPVPNGLD-AGPAALTEPMAVGLHAVNT--SRIERNETALVIGCGP
MAB_0526c|M.abscessus_ATCC_199 P-LLLPIPNGLD-PRHAALTEPMAVGLHAVNK--SAIQPGTGAIVIGCGP
MMAR_3328|M.marinum_M P-LLIEVPDHVP-TEVAALTEPLAVAYRTVAR--VDTTTDDVALVVGCGP
Mflv_0660|M.gilvum_PYR-GCK R-LAVRVPDVVD-LEEAALTEPTAVAFHAVRR--AELLLGETVMVLGAGA
TH_4349|M.thermoresistible__bu ERLVVKLPESMTPVTAAALTDAGLTPYHAIRRSWPKLTPTSTAVVIGVGG
MUL_4777|M.ulcerans_Agy99 A-GLTRVPDNVSLDQAAILADAVSTPFGAVVR-TGKVAVGESVGVWGVGG
MLBr_01784|M.leprae_Br4923 G-QCTKVDPAADPAVAGLLGCGVMAGLGAAIN-TAAVSRDDTVAVIGCGG
: . * . : . * * *
Mb3753|M.bovis_AF2122/97 IGLAVICMLKSRGVHTVIASDFSPGRRALATACGADSVVDPVQDSPYAVA
Rv3726|M.tuberculosis_H37Rv IGLAVICMLKSRGVHTVIASDFSPGRRALATACGADSVVDPVQDSPYAVA
Mvan_5762|M.vanbaalenii_PYR-1 VGLAVICHLKALGVRTIVASDFSPGRRALAARCGADVVVDPAVDSPYEAV
MAV_0537|M.avium_104 IGIAIIAALATRGVESIVASDFSPKRRQLATAMGAHRTLDPAQGSPFDSV
MSMEG_6108|M.smegmatis_MC2_155 VGIAVIAALKLLGVEDIVASDFSPTRRRLAAAMGAHVTLDPAQDSPFHQC
MAB_0526c|M.abscessus_ATCC_199 VGIAVIAALHNLGIEPIVASDYSSARRDLAQRMGAHQVVDPKIEPTFHAW
MMAR_3328|M.marinum_M IGLAVIAVLKMKGVGPIVASDFSPERRALATQLGADVVLDPNENSPFEAF
Mflv_0660|M.gilvum_PYR-GCK LGLTVIQCARAAGAARIFVTEPSGVRASLARDLGATLVLDPHDPGTTACI
TH_4349|M.thermoresistible__bu LGHVAVQLVKATTAARLIAVDNRSEALELAREMGADHVVRSDEQAVEAIW
MUL_4777|M.ulcerans_Agy99 VGTHIVQLARLVGAVPVIALDINPVVLDRALELGADYAFDSRDDQLHDKL
MLBr_01784|M.leprae_Br4923 VGDAAISGAALVGANRIIAVDIDDTKLEWARTFGATHTVNALELEVVKTI
:* : :.. : * ** .. .
Mb3753|M.bovis_AF2122/97 AGLGQGNRHLQSILDAFDLAVGTVESLQRLRLPWWHLWRAAEAAGAATPK
Rv3726|M.tuberculosis_H37Rv AGLGQGNRHLQSILDAFDLAVGTVERLQRLRLPWWHLWRAAEAAGAATPK
Mvan_5762|M.vanbaalenii_PYR-1 PAKQRG---LTELPALYELGVGSMEKLRKIPG-WSHVYRVADRLGGTAPK
MAV_0537|M.avium_104 K-------------------------------------------------
MSMEG_6108|M.smegmatis_MC2_155 R-------------------------------------------------
MAB_0526c|M.abscessus_ATCC_199 SRIGAG--------------------------------------------
MMAR_3328|M.marinum_M MAAAVTDDPARMAP-------------------------AHTVFGQLPLR
Mflv_0660|M.gilvum_PYR-GCK LEETRG------------------------------------------VG
TH_4349|M.thermoresistible__bu ELTGGR-------------------------------------------G
MUL_4777|M.ulcerans_Agy99 AEITGG------------------------------------------RM
MLBr_01784|M.leprae_Br4923 QDLTSG------------------------------------------FG
Mb3753|M.bovis_AF2122/97 RPVIFECVGVPGIIDGIIASAPLFSRVVVVGVCMGSD----HIRPAMAIN
Rv3726|M.tuberculosis_H37Rv RPVIFECVGVPGIIDGIIASAPLFSRVVVVGVCMGSD----HIRPAMAIN
Mvan_5762|M.vanbaalenii_PYR-1 RPVIFECVGVPGMIDGIIGAAPLASRVVVVGVCMGDD----KLRPAMAIG
MAV_0537|M.avium_104 PAVVFEAVGVPGIIDDVLLRARPGTRLVVAGVCMQPD----TVHPFFAIA
MSMEG_6108|M.smegmatis_MC2_155 PAVVFEAVGAPGIIDDILRRCPAGTRVVVAGVCMQPD----TVHPLYAAV
MAB_0526c|M.abscessus_ATCC_199 APVIFEAVGVPGILNEVLRDAPRSARVVVVGVCMEPD----AITPYFGIA
MMAR_3328|M.marinum_M PTVAFECVGVPGLIQQLIAGVPPGTRIGVAGINMSSD----HFEPAQCIA
Mflv_0660|M.gilvum_PYR-GCK VDVVFHVAGSAEAFTQGLDCLRKQGRFMEMSSWAGAA----SLDVNRHLL
TH_4349|M.thermoresistible__bu ADVLLDFVGADATIELARAAARPLGDVTIVGIAGGSV----PLS-FFSQP
MUL_4777|M.ulcerans_Agy99 LDVAFDAVGLKATFEQALGSLTAGGRLVGVGTSAESP----TVGPTSMFG
MLBr_01784|M.leprae_Br4923 VDVVIDTVGRPETWKQAFYARDLAGTVVLVGVPTPDMRLDMPLLDFFSHG
* :. .* . . .
Mb3753|M.bovis_AF2122/97 KEINLRFVLGYTP-LEFRDTLHMLADGKVNAAPLITGTVGLP-GVAAAFD
Rv3726|M.tuberculosis_H37Rv KEINLRFVLGYTP-LEFRDTLHMLADGKVNAAPLITGTVGLP-GVAAAFD
Mvan_5762|M.vanbaalenii_PYR-1 KEIDLRFVFGYTP-LEFRDTLHMLADGKLDASALVTGTVGLG-GVAAAFD
MAV_0537|M.avium_104 KEINVQFVLAYTP-EEFGDSLRALAEGDIDVAPLITGEVGLD-GVGAAFD
MSMEG_6108|M.smegmatis_MC2_155 KQISVSFVFGYDP-AEFASSLRAIAEGDIDVRPMITGSVPLD-GMADAFD
MAB_0526c|M.abscessus_ATCC_199 KEVAVQFVLAYEP-AEFSETLRRIAHGEIDVAPLVTGEVGLE-DVGSAFA
MMAR_3328|M.marinum_M KELDIRFSLYYTL-EEFAQTFAHLAAGDFDVAPLLTGTVALE-GVADAFE
Mflv_0660|M.gilvum_PYR-GCK KEIQLRMVFGYDMFDDFPAVLALIADGKLALAPQITARVPLDRAVKEGLG
TH_4349|M.thermoresistible__bu YEVSIQTTYWGTR-PELVELLDLAARG---LVHVESTTYPLD-DALQAYR
MUL_4777|M.ulcerans_Agy99 LTHKQVLGHLGYQNVDIETLAKLVSLGRLDLSRSISEIVPLE-DLSLGIE
MLBr_01784|M.leprae_Br4923 GSLKSSWYGDCLPERDFPTLVDLYLQGRLPLGKFVSERIGLG-DVEEAFH
:: * : * .
Mb3753|M.bovis_AF2122/97 ALGD-PEAHAKIMIDPKSNAASPQPFRVE
Rv3726|M.tuberculosis_H37Rv ALGD-PEAHAKIMIDPKSNAASPQPFRVE
Mvan_5762|M.vanbaalenii_PYR-1 ALGD-PETHAKILIDPAGTAVTP------
MAV_0537|M.avium_104 DLAD-PERHCKVLVTP-------------
MSMEG_6108|M.smegmatis_MC2_155 ELAT-PDAHCKILVTP-------------
MAB_0526c|M.abscessus_ATCC_199 DLGD-PERHCKILVVP-------------
MMAR_3328|M.marinum_M RLRN-NPHDAKILLDPTR-----------
Mflv_0660|M.gilvum_PYR-GCK GLLEGREGLVKVLVKP-------------
TH_4349|M.thermoresistible__bu DLRAGRVRGRAVIVP--------------
MUL_4777|M.ulcerans_Agy99 KLERQDGNPIRILVQP-------------
MLBr_01784|M.leprae_Br4923 KIHGGKVLRSVVML---------------
: :::