For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MKAVSCVHGTLEVVDLPSPRPGEGQLVLDVLRCGICGSDLHAKDHADELTEVMAEGGYHDFVRSDTPVVM GHEFCGVVADRGRKVGKEFKPGSTVVAFPLMRAHGGVHLTGLSPLAPGGYAEQVVAEAALTFVVPNGLDP ATAALTEPMAVAYHAVRRSDVTTKDVAIVLGCGPVGLAVICHLKALGVRTIVASDFSPGRRALAARCGAD VVVDPAVDSPYEAVPAKQRGLTELPALYELGVGSMEKLRKIPGWSHVYRVADRLGGTAPKRPVIFECVGV PGMIDGIIGAAPLASRVVVVGVCMGDDKLRPAMAIGKEIDLRFVFGYTPLEFRDTLHMLADGKLDASALV TGTVGLGGVAAAFDALGDPETHAKILIDPAGTAVTP
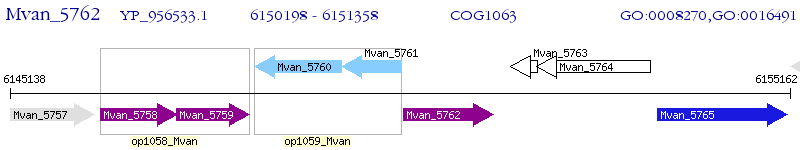
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5762 | - | - | 100% (386) | alcohol dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0527 | - | 3e-27 | 26.94% (386) | alcohol dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0490 | - | 1e-25 | 28.17% (394) | alcohol dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3753 | - | 1e-142 | 64.45% (391) | putative dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0638 | - | 2e-27 | 26.94% (386) | alcohol dehydrogenase |
| M. tuberculosis H37Rv | Rv3726 | - | 1e-142 | 64.45% (391) | dehydrogenase |
| M. leprae Br4923 | MLBr_01784 | adhE2 | 3e-13 | 24.74% (384) | putative alcohol dehydrogenase (Zn dependent) |
| M. abscessus ATCC 19977 | MAB_0526c | - | 5e-73 | 43.09% (369) | hypothetical protein MAB_0526c |
| M. marinum M | MMAR_3328 | adhE2_1 | 1e-78 | 45.00% (380) | zinc-dependent alcohol dehydrogenase AdhE2_1 |
| M. avium 104 | MAV_0537 | - | 3e-72 | 41.30% (368) | zinc-binding dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6108 | - | 1e-71 | 41.19% (369) | zinc-binding dehydrogenase |
| M. thermoresistible (build 8) | TH_4627 | - | 1e-16 | 25.63% (398) | PUTATIVE Alcohol dehydrogenase, zinc-binding domain protein |
| M. ulcerans Agy99 | MUL_1292 | adhE2 | 2e-16 | 25.58% (387) | zinc-dependent alcohol dehydrogenase AdhE2 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3753|M.bovis_AF2122/97 ----MKAVTCTN--AKLEVVDRPSPAPAKGQLLLDVLRCGICGSDLHARL
Rv3726|M.tuberculosis_H37Rv ----MKAVTCTN--AKLEVVDRPSPAPAKGQLLLDVLRCGICGSDLHARL
Mvan_5762|M.vanbaalenii_PYR-1 ----MKAVSCVH--GTLEVVDLPSPRPGEGQLVLDVLRCGICGSDLHAKD
MAV_0537|M.avium_104 ----MRAAVLRDG-RMVYRDDVPEPIPETGQVLVGVRACGICGSDLHFAA
MSMEG_6108|M.smegmatis_MC2_155 ----MRAAVLRQG-RMVVRDDVAEPVPGPGQVLVEVKACGICGSDLHFAQ
MAB_0526c|M.abscessus_ATCC_199 ----MRASVLRNG-SMVYRDDVPSPTPGEGQVLVKVSACGICGSDLHFAK
MMAR_3328|M.marinum_M ----MRAVLMQD--SQLQVAEVPEPVPGPGEVLVDVLACGICGSDLHCAA
MLBr_01784|M.leprae_Br4923 MSQTVRGVISRKKDEPVELVDIVIPDPGPGEAVVDVTACGVCHTDLTYRE
MUL_1292|M.ulcerans_Agy99 MSQTVRGVISRKKGEPVELVDIVVPDPGPGEAVVDISACGVCHTDLTYRE
TH_4627|M.thermoresistible__bu MSQSVRGVIARKKDEPVEVVDIVIPDPGPGEVVVKVTACGVCHTDLHYRE
Mflv_0638|M.gilvum_PYR-GCK ----MRAAIYHGR-EDVRIEELPDPSPRAGEVVIEVARAGICGTDLHEYI
::. : : * * *: :: : .*:* :**
Mb3753|M.bovis_AF2122/97 HC-DELADVMAESGY----HAFMRSNQQVVFGHEFCGEVVDYGPGTRRTP
Rv3726|M.tuberculosis_H37Rv HC-DELADVMAESGY----HAFMRSNQQVVFGHEFCGEVVDYGPGTRRTP
Mvan_5762|M.vanbaalenii_PYR-1 HA-DELTEVMAEGGY----HDFVRSDTPVVMGHEFCGVVADRGRKVGKEF
MAV_0537|M.avium_104 HG-AQVLQMSDRVAGGAAGMR-VDLDHDIFMGHEFSAEILEAGPDTETHP
MSMEG_6108|M.smegmatis_MC2_155 HG-EDLLASTAQLEGVANQNDDVDLSRDIFMGHEFSAEILEAGPSTDTYA
MAB_0526c|M.abscessus_ATCC_199 HG-ARAIELSKQMVGMPSLTDGVDLSTDVFMGHEFAAEVIEAGPRTQAPA
MMAR_3328|M.marinum_M HG-PEFNSATQAAGG----FDMMDLSRPVVFGHEFVGRIAAYGPDTVQRI
MLBr_01784|M.leprae_Br4923 GG-INDRYPFLLGHEAAGTVEVVGPGVTAVEPGDFVILNWRAVCGQCRAC
MUL_1292|M.ulcerans_Agy99 GG-INDEYPFLLGHEAAGTVEAVGPGVTNVEPGDFVILNWRAVCGQCRAC
TH_4627|M.thermoresistible__bu GG-INDEFPFLLGHEAAGTVESVGSGVTNVEPGDFVVLNWRAVCGQCRAC
Mflv_0638|M.gilvum_PYR-GCK AGPMHAAPGVVIGHEYSGTVVGVGSGVREFTEGDRVCGVGVFGCGECGFC
: . . :
Mb3753|M.bovis_AF2122/97 RRGTPVVAMPLLRRGNKE---VHGIGLSTMAPGAYAERLVVEQSLTFPVP
Rv3726|M.tuberculosis_H37Rv RRGTPVVAMPLLRRGNKE---VHGIGLSTMAPGAYAERLVVEQSLTFPVP
Mvan_5762|M.vanbaalenii_PYR-1 KPGSTVVAFPLMR-AHGG---VHLTGLSPLAPGGYAEQVVAEAALTFVVP
MAV_0537|M.avium_104 P-GTLVTSLPVLLSAKG----VEPIVYSNTTIGGYAERMLLSAPLLLPIP
MSMEG_6108|M.smegmatis_MC2_155 P-GTLVTSVPVLISPQG----MDMIVYSNTTLGGYAERMLLSAPLLLPVP
MAB_0526c|M.abscessus_ATCC_199 S-GTAVTSIPILLSAKG----LEPIVYSNTTLGGYAEQMLLSAPLLLPIP
MMAR_3328|M.marinum_M PVGRRVVSAPFLLREQK----VTLGFAGPEAPGGYSERMLLSEPLLIEVP
MLBr_01784|M.leprae_Br4923 KRGRPSYCFDTFNAQQKMTLIDGTELTPALGIGAFADKTLVHSGQCTKVD
MUL_1292|M.ulcerans_Agy99 KRGRPSYCFDTFNAEQKMTLTDGTELTPALGIGAFADKTLVHSGQCTKVD
TH_4627|M.thermoresistible__bu KRGRPHLCFDTFNATQKMTLGDGTELTPALGIGAFAEKTLVHEGQCTKVD
Mflv_0638|M.gilvum_PYR-GCK KQGAEALCG-------------AVGFIGFAVNGALARYASLPTKALFRIP
* . *. : :
Mb3753|M.bovis_AF2122/97 NGLAPEIAALTEP--MAVGWHAVRRGEVGKGDVAIVIGCGPIGLAVICML
Rv3726|M.tuberculosis_H37Rv NGLAPEIAALTEP--MAVGWHAVRRGEVGKGDVAIVIGCGPIGLAVICML
Mvan_5762|M.vanbaalenii_PYR-1 NGLDPATAALTEP--MAVAYHAVRRSDVTTKDVAIVLGCGPVGLAVICHL
MAV_0537|M.avium_104 NGLDVKHAALTEP--MAVGLHAVNKSNITPAETALVLGCGPIGIAIIAAL
MSMEG_6108|M.smegmatis_MC2_155 NGLDAGPAALTEP--MAVGLHAVNTSRIERNETALVIGCGPVGIAVIAAL
MAB_0526c|M.abscessus_ATCC_199 NGLDPRHAALTEP--MAVGLHAVNKSAIQPGTGAIVIGCGPVGIAVIAAL
MMAR_3328|M.marinum_M DHVPTEVAALTEP--LAVAYRTVARVDTTTDDVALVVGCGPIGLAVIAVL
MLBr_01784|M.leprae_Br4923 PAADPAVAGLLGCGVMAGLGAAINTAAVSRDDTVAVIGCGGVGDAAISGA
MUL_1292|M.ulcerans_Agy99 PAADPAVAGLLGCGVMAGLGAAVNTGGVTRDDTVAVIGCGGVGDAAIAGA
TH_4627|M.thermoresistible__bu PAADPAVAGLLGCGVMAGVGAAINTGAVSRDDTVAVIGCGGVGDAAIAGA
Mflv_0638|M.gilvum_PYR-GCK DEISLAEAAVVEP--IASAYHAVRRSGLAAGGTVFIAGAGPIGLALVQFS
*.: :* :: . : *.* :* * :
Mb3753|M.bovis_AF2122/97 KSRGVHTVIASDFSPGRRALATACGADSVVDPVQDSPYAVAAGLGQGNRH
Rv3726|M.tuberculosis_H37Rv KSRGVHTVIASDFSPGRRALATACGADSVVDPVQDSPYAVAAGLGQGNRH
Mvan_5762|M.vanbaalenii_PYR-1 KALGVRTIVASDFSPGRRALAARCGADVVVDPAVDSPYEAVPAKQRG---
MAV_0537|M.avium_104 ATRGVESIVASDFSPKRRQLATAMGAHRTLDPAQGSPFDSVK--------
MSMEG_6108|M.smegmatis_MC2_155 KLLGVEDIVASDFSPTRRRLAAAMGAHVTLDPAQDSPFHQCR--------
MAB_0526c|M.abscessus_ATCC_199 HNLGIEPIVASDYSSARRDLAQRMGAHQVVDPKIEPTFHAWSRIGA----
MMAR_3328|M.marinum_M KMKGVGPIVASDFSPERRALATQLGADVVLDPNENSPFEAFMAAAVTDDP
MLBr_01784|M.leprae_Br4923 ALVGANRIIAVDIDDTKLEWARTFGATHTVNALELEVVKTIQDLTSG---
MUL_1292|M.ulcerans_Agy99 ALVGAKKIIAVDTDNTKLEWARAFGATHTVNARELDVVETIQDLTGG---
TH_4627|M.thermoresistible__bu ALVGAKKIIAVDTDAKKLDWAREFGATHTIDASSTDVVEAIQELTDG---
Mflv_0638|M.gilvum_PYR-GCK LAQGATQVIVSEVSATRRVAAHRVGATRVIDPLAEDAVEVVRTLTNG---
* ::. : . : * ** .::.
Mb3753|M.bovis_AF2122/97 LQSILDAFDLAVGTVESLQRLRLPWWHLWRAAEAAGAATPKRPVIFECVG
Rv3726|M.tuberculosis_H37Rv LQSILDAFDLAVGTVERLQRLRLPWWHLWRAAEAAGAATPKRPVIFECVG
Mvan_5762|M.vanbaalenii_PYR-1 LTELPALYELGVGSMEKLRKIPG-WSHVYRVADRLGGTAPKRPVIFECVG
MAV_0537|M.avium_104 -----------------------------------------PAVVFEAVG
MSMEG_6108|M.smegmatis_MC2_155 -----------------------------------------PAVVFEAVG
MAB_0526c|M.abscessus_ATCC_199 ----------------------------------------GAPVIFEAVG
MMAR_3328|M.marinum_M ARMAPAHTVFGQ-------------------------LPLRPTVAFECVG
MLBr_01784|M.leprae_Br4923 ---------------------------------------FGVDVVIDTVG
MUL_1292|M.ulcerans_Agy99 ---------------------------------------FGVDVVIDAVG
TH_4627|M.thermoresistible__bu ---------------------------------------FGADVVVDAVG
Mflv_0638|M.gilvum_PYR-GCK ---------------------------------------NGVDISFDAAG
: .: .*
Mb3753|M.bovis_AF2122/97 VPGIIDGIIASAPLFSRVVVVGVCMGSDHIRP----AMAINKEINLRFVL
Rv3726|M.tuberculosis_H37Rv VPGIIDGIIASAPLFSRVVVVGVCMGSDHIRP----AMAINKEINLRFVL
Mvan_5762|M.vanbaalenii_PYR-1 VPGMIDGIIGAAPLASRVVVVGVCMGDDKLRP----AMAIGKEIDLRFVF
MAV_0537|M.avium_104 VPGIIDDVLLRARPGTRLVVAGVCMQPDTVHP----FFAIAKEINVQFVL
MSMEG_6108|M.smegmatis_MC2_155 APGIIDDILRRCPAGTRVVVAGVCMQPDTVHP----LYAAVKQISVSFVF
MAB_0526c|M.abscessus_ATCC_199 VPGILNEVLRDAPRSARVVVVGVCMEPDAITP----YFGIAKEVAVQFVL
MMAR_3328|M.marinum_M VPGLIQQLIAGVPPGTRIGVAGINMSSDHFEP----AQCIAKELDIRFSL
MLBr_01784|M.leprae_Br4923 RPETWKQAFYARDLAGTVVLVGVPTPDMRLDMPLLDFFSHGGSLKSSWYG
MUL_1292|M.ulcerans_Agy99 RPETWKQAFYARDLAGTVVLVGVPTPDMRLDMPLVDFFSRGGALKSSWYG
TH_4627|M.thermoresistible__bu RPETWEQAFYARDLAGTVVLVGVPTPDMRIDMPLIDFFSRGGSLKSSWYG
Mflv_0638|M.gilvum_PYR-GCK VQPALDAALGVLRPRGRLMVVAIWEAPAGIDIN----RSVMREANIGFSF
. : : :..: . :
Mb3753|M.bovis_AF2122/97 GYTP-LEFRDTLHMLADGKVNAAPLITGTVGLPGVAAA-FDALGD-PEAH
Rv3726|M.tuberculosis_H37Rv GYTP-LEFRDTLHMLADGKVNAAPLITGTVGLPGVAAA-FDALGD-PEAH
Mvan_5762|M.vanbaalenii_PYR-1 GYTP-LEFRDTLHMLADGKLDASALVTGTVGLGGVAAA-FDALGD-PETH
MAV_0537|M.avium_104 AYTP-EEFGDSLRALAEGDIDVAPLITGEVGLDGVGAA-FDDLAD-PERH
MSMEG_6108|M.smegmatis_MC2_155 GYDP-AEFASSLRAIAEGDIDVRPMITGSVPLDGMADA-FDELAT-PDAH
MAB_0526c|M.abscessus_ATCC_199 AYEP-AEFSETLRRIAHGEIDVAPLVTGEVGLEDVGSA-FADLGD-PERH
MMAR_3328|M.marinum_M YYTL-EEFAQTFAHLAAGDFDVAPLLTGTVALEGVADA-FERLRN-NPHD
MLBr_01784|M.leprae_Br4923 DCLPERDFPTLVDLYLQGRLPLGKFVSERIGLGDVEEA-FHKIHG-GKVL
MUL_1292|M.ulcerans_Agy99 DCLPERDFPTLIDLYLQGRLPLDKFVSEQIGLGGIEEA-FEKMHA-GKVL
TH_4627|M.thermoresistible__bu DCLPERDFPTLINLYLQGRLPLEKFVTERIALDEVEGA-FEKMQR-GEVL
Mflv_0638|M.gilvum_PYR-GCK CYEAQRQVPAILDLLATGAINLGELITDEIPLDAVVSQGLEELRVNRDAH
:. . * . ::: : * : : :
Mb3753|M.bovis_AF2122/97 AKIMIDPKSNAASPQPFRVE
Rv3726|M.tuberculosis_H37Rv AKIMIDPKSNAASPQPFRVE
Mvan_5762|M.vanbaalenii_PYR-1 AKILIDPAGTAVTP------
MAV_0537|M.avium_104 CKVLVTP-------------
MSMEG_6108|M.smegmatis_MC2_155 CKILVTP-------------
MAB_0526c|M.abscessus_ATCC_199 CKILVVP-------------
MMAR_3328|M.marinum_M AKILLDPTR-----------
MLBr_01784|M.leprae_Br4923 RSVVML--------------
MUL_1292|M.ulcerans_Agy99 RSVVVL--------------
TH_4627|M.thermoresistible__bu RSVVVLK-------------
Mflv_0638|M.gilvum_PYR-GCK VKILIDPSA-----------
.:::