For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRAAVLRQGRMVVRDDVAEPVPGPGQVLVEVKACGICGSDLHFAQHGEDLLASTAQLEGVANQNDDVDLS RDIFMGHEFSAEILEAGPSTDTYAPGTLVTSVPVLISPQGMDMIVYSNTTLGGYAERMLLSAPLLLPVPN GLDAGPAALTEPMAVGLHAVNTSRIERNETALVIGCGPVGIAVIAALKLLGVEDIVASDFSPTRRRLAAA MGAHVTLDPAQDSPFHQCRPAVVFEAVGAPGIIDDILRRCPAGTRVVVAGVCMQPDTVHPLYAAVKQISV SFVFGYDPAEFASSLRAIAEGDIDVRPMITGSVPLDGMADAFDELATPDAHCKILVTP
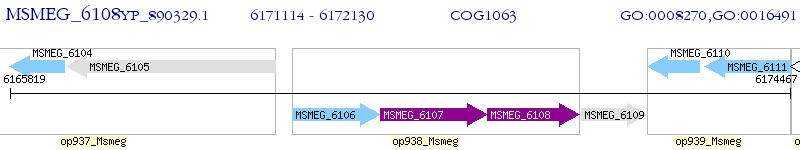
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6108 | - | - | 100% (338) | zinc-binding dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_3605 | - | 1e-29 | 27.45% (357) | sorbitol dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2914 | - | 8e-28 | 29.10% (323) | L-idonate 5-dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3753 | - | 7e-69 | 39.33% (389) | putative dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0638 | - | 5e-27 | 26.11% (360) | alcohol dehydrogenase |
| M. tuberculosis H37Rv | Rv3726 | - | 8e-69 | 39.33% (389) | dehydrogenase |
| M. leprae Br4923 | MLBr_01784 | adhE2 | 3e-15 | 26.20% (229) | putative alcohol dehydrogenase (Zn dependent) |
| M. abscessus ATCC 19977 | MAB_0526c | - | 1e-116 | 59.77% (343) | hypothetical protein MAB_0526c |
| M. marinum M | MMAR_5116 | - | 1e-128 | 65.98% (341) | dehydrogenase |
| M. avium 104 | MAV_0537 | - | 1e-135 | 71.30% (338) | zinc-binding dehydrogenase |
| M. thermoresistible (build 8) | TH_4631 | - | 2e-19 | 28.99% (207) | PUTATIVE Alcohol dehydrogenase GroES domain protein |
| M. ulcerans Agy99 | MUL_4777 | - | 8e-21 | 26.06% (353) | zinc-dependent alcohol dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_5762 | - | 8e-72 | 41.19% (369) | alcohol dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_5116|M.marinum_M -----MRASVLRNGHMTYRDDVAEPTPGQGQVLVGVRACGICGSDLHFAA
MAV_0537|M.avium_104 -----MRAAVLRDGRMVYRDDVPEPIPETGQVLVGVRACGICGSDLHFAA
MSMEG_6108|M.smegmatis_MC2_155 -----MRAAVLRQGRMVVRDDVAEPVPGPGQVLVEVKACGICGSDLHFAQ
MAB_0526c|M.abscessus_ATCC_199 -----MRASVLRNGSMVYRDDVPSPTPGEGQVLVKVSACGICGSDLHFAK
Mb3753|M.bovis_AF2122/97 -----MKAVTCTNAKLEVVD-RPSPAPAKGQLLLDVLRCGICGSDLHARL
Rv3726|M.tuberculosis_H37Rv -----MKAVTCTNAKLEVVD-RPSPAPAKGQLLLDVLRCGICGSDLHARL
Mvan_5762|M.vanbaalenii_PYR-1 -----MKAVSCVHGTLEVVD-LPSPRPGEGQLVLDVLRCGICGSDLHAKD
Mflv_0638|M.gilvum_PYR-GCK -----MRAAIYHGREDVRIEELPDPSPRAGEVVIEVARAGICGTDLHEYI
TH_4631|M.thermoresistible__bu ----------------------------------------VNGXXXEETR
MUL_4777|M.ulcerans_Agy99 MAATMRAERFYADSKTVVLEDVPIPEPGPGEVLVKVAYCGICHSDLSLIN
MLBr_01784|M.leprae_Br4923 MSQTVRGVISRKKDEPVELVDIVIPDPGPGEAVVDVTACGVCHTDLTYRE
:
MMAR_5116|M.marinum_M HGPEVLAMSSRLSHGDGYTN-TDLSRDVFMGHEFCAEVIEAGPDT-DTYP
MAV_0537|M.avium_104 HGAQVLQMSDRVAGGAAGMR-VDLDHDIFMGHEFSAEILEAGPDT-ETHP
MSMEG_6108|M.smegmatis_MC2_155 HGEDLLASTAQLEGVANQNDDVDLSRDIFMGHEFSAEILEAGPST-DTYA
MAB_0526c|M.abscessus_ATCC_199 HGARAIELSKQMVGMPSLTDGVDLSTDVFMGHEFAAEVIEAGPRT-QAPA
Mb3753|M.bovis_AF2122/97 HCDELADVMAESG----YHAFMRSNQQVVFGHEFCGEVVDYGPGTRRTPR
Rv3726|M.tuberculosis_H37Rv HCDELADVMAESG----YHAFMRSNQQVVFGHEFCGEVVDYGPGTRRTPR
Mvan_5762|M.vanbaalenii_PYR-1 HADELTEVMAEGG----YHDFVRSDTPVVMGHEFCGVVADRGRKVGKEFK
Mflv_0638|M.gilvum_PYR-GCK AGP------------------MHAAPGVVIGHEYSGTVVGVGSGV-REFT
TH_4631|M.thermoresistible__bu RGR----------------------------YNLCPHMRFYGTPP-----
MUL_4777|M.ulcerans_Agy99 GTFP------------------TQAPVVTQGHEASGTIVKLGPEV-DGWS
MLBr_01784|M.leprae_Br4923 GGIN-------------------DRYPFLLGHEAAGTVEVVGPGV-TAVE
:: . : *
MMAR_5116|M.marinum_M AGTLVTSIPVLL------SAKGVEPIFSSN------------------DT
MAV_0537|M.avium_104 PGTLVTSLPVLL------SAKGVEPIVYSN------------------TT
MSMEG_6108|M.smegmatis_MC2_155 PGTLVTSVPVLI------SPQGMDMIVYSN------------------TT
MAB_0526c|M.abscessus_ATCC_199 SGTAVTSIPILL------SAKGLEPIVYSN------------------TT
Mb3753|M.bovis_AF2122/97 RGTPVVAMPLLRR-----GNKEVHGIGLST------------------MA
Rv3726|M.tuberculosis_H37Rv RGTPVVAMPLLRR-----GNKEVHGIGLST------------------MA
Mvan_5762|M.vanbaalenii_PYR-1 PGSTVVAFPLMR------AHGGVHLTGLSP------------------LA
Mflv_0638|M.gilvum_PYR-GCK EGDRVCGVGVFGCGECGFCKQGAEALCGAVG-------------FIGFAV
TH_4631|M.thermoresistible__bu -------------------------------------------------V
MUL_4777|M.ulcerans_Agy99 EGDRVVVAAGRPCQSCPNCRRGDTVNCLRIQ-------------LMAFAY
MLBr_01784|M.leprae_Br4923 PGDFVILNWRAVCGQCRACKRGRPSYCFDTFNAQQKMTLIDGTELTPALG
MMAR_5116|M.marinum_M IGGYAERMLLSAPLLLPVPNGLDFRHAALTEPMAVG--LHAVNKAGVDVG
MAV_0537|M.avium_104 IGGYAERMLLSAPLLLPIPNGLDVKHAALTEPMAVG--LHAVNKSNITPA
MSMEG_6108|M.smegmatis_MC2_155 LGGYAERMLLSAPLLLPVPNGLDAGPAALTEPMAVG--LHAVNTSRIERN
MAB_0526c|M.abscessus_ATCC_199 LGGYAEQMLLSAPLLLPIPNGLDPRHAALTEPMAVG--LHAVNKSAIQPG
Mb3753|M.bovis_AF2122/97 PGAYAERLVVEQSLTFPVPNGLAPEIAALTEPMAVG--WHAVRRGEVGKG
Rv3726|M.tuberculosis_H37Rv PGAYAERLVVEQSLTFPVPNGLAPEIAALTEPMAVG--WHAVRRGEVGKG
Mvan_5762|M.vanbaalenii_PYR-1 PGGYAEQVVAEAALTFVVPNGLDPATAALTEPMAVA--YHAVRRSDVTTK
Mflv_0638|M.gilvum_PYR-GCK NGALARYASLPTKALFRIPDEISLAEAAVVEPIASA--YHAVRRSGLAAG
TH_4631|M.thermoresistible__bu DGALCEYVTIGAAFAHPVPAEMTDNAAALCEPLSVA--IATVDKAAVAGG
MUL_4777|M.ulcerans_Agy99 DGAWAEYTVAQAAGLTRVPDNVSLDQAAILADAVSTPFGAVVRTGKVAVG
MLBr_01784|M.leprae_Br4923 IGAFADKTLVHSGQCTKVDPAADPAVAGLLGCGVMAGLGAAINTAAVSRD
*. . : *.: .: . :
MMAR_5116|M.marinum_M DTALVLGCGPIGIAIIAALRMRGVETIVASDFSQKRRELASIMGAHQTVD
MAV_0537|M.avium_104 ETALVLGCGPIGIAIIAALATRGVESIVASDFSPKRRQLATAMGAHRTLD
MSMEG_6108|M.smegmatis_MC2_155 ETALVIGCGPVGIAVIAALKLLGVEDIVASDFSPTRRRLAAAMGAHVTLD
MAB_0526c|M.abscessus_ATCC_199 TGAIVIGCGPVGIAVIAALHNLGIEPIVASDYSSARRDLAQRMGAHQVVD
Mb3753|M.bovis_AF2122/97 DVAIVIGCGPIGLAVICMLKSRGVHTVIASDFSPGRRALATACGADSVVD
Rv3726|M.tuberculosis_H37Rv DVAIVIGCGPIGLAVICMLKSRGVHTVIASDFSPGRRALATACGADSVVD
Mvan_5762|M.vanbaalenii_PYR-1 DVAIVLGCGPVGLAVICHLKALGVRTIVASDFSPGRRALAARCGADVVVD
Mflv_0638|M.gilvum_PYR-GCK GTVFIAGAGPIGLALVQFSLAQGATQVIVSEVSATRRVAAHRVGATRVID
TH_4631|M.thermoresistible__bu SRVLIAGAGPIGLMTAQVARAYGATDIVVTDLDPHRRRLAHRFGATTTLD
MUL_4777|M.ulcerans_Agy99 ESVGVWGVGGVGTHIVQLARLVGAVPVIALDINPVVLDRALELGADYAFD
MLBr_01784|M.leprae_Br4923 DTVAVIGCGGVGDAAISGAALVGANRIIAVDIDDTKLEWARTFGATHTVN
. : * * :* * ::. : . * ** ..:
MMAR_5116|M.marinum_M PAVSSPFEHVK---------------------------------------
MAV_0537|M.avium_104 PAQGSPFDSVK---------------------------------------
MSMEG_6108|M.smegmatis_MC2_155 PAQDSPFHQCR---------------------------------------
MAB_0526c|M.abscessus_ATCC_199 PKIEPTFHAWSR--------------------------------------
Mb3753|M.bovis_AF2122/97 PVQDSPYAVAAGLGQGNRHLQSILDAFDLAVGTVESLQRLRLPWWHLWRA
Rv3726|M.tuberculosis_H37Rv PVQDSPYAVAAGLGQGNRHLQSILDAFDLAVGTVERLQRLRLPWWHLWRA
Mvan_5762|M.vanbaalenii_PYR-1 PAVDSPYEAVPAKQRG---LTELPALYELGVGSMEKLRKIPG-WSHVYRV
Mflv_0638|M.gilvum_PYR-GCK PLAEDAVEVVRTLTNG----------------------------------
TH_4631|M.thermoresistible__bu PQTDDVTGLRVD--------------------------------------
MUL_4777|M.ulcerans_Agy99 SRDDQLHDKLAEITGG----------------------------------
MLBr_01784|M.leprae_Br4923 ALELEVVKTIQDLTSG----------------------------------
.
MMAR_5116|M.marinum_M ----------ATVVFEAVGVPGMIDEVILRARPGTRLIVAGVCMQPDTVH
MAV_0537|M.avium_104 ----------PAVVFEAVGVPGIIDDVLLRARPGTRLVVAGVCMQPDTVH
MSMEG_6108|M.smegmatis_MC2_155 ----------PAVVFEAVGAPGIIDDILRRCPAGTRVVVAGVCMQPDTVH
MAB_0526c|M.abscessus_ATCC_199 ------IGAGAPVIFEAVGVPGILNEVLRDAPRSARVVVVGVCMEPDAIT
Mb3753|M.bovis_AF2122/97 AEAAGAATPKRPVIFECVGVPGIIDGIIASAPLFSRVVVVGVCMGSDHIR
Rv3726|M.tuberculosis_H37Rv AEAAGAATPKRPVIFECVGVPGIIDGIIASAPLFSRVVVVGVCMGSDHIR
Mvan_5762|M.vanbaalenii_PYR-1 ADRLGGTAPKRPVIFECVGVPGMIDGIIGAAPLASRVVVVGVCMGDDKLR
Mflv_0638|M.gilvum_PYR-GCK --------NGVDISFDAAGVQPALDAALGVLRPRGRLMVVAIWEAPAGID
TH_4631|M.thermoresistible__bu ------------AFIDASGAPAAVMSGLAAVRPAGRAVLVGMGAETMELP
MUL_4777|M.ulcerans_Agy99 --------RMLDVAFDAVGLKATFEQALGSLTAGGRLVGVGTSAESPTVG
MLBr_01784|M.leprae_Br4923 --------FGVDVVIDTVGRPETWKQAFYARDLAGTVVLVGVPTPDMRLD
:: * : : .. :
MMAR_5116|M.marinum_M PFFAI-TKEINVQFVLAYDPE----EFATSLRALAEGLIDVSPLITGEVD
MAV_0537|M.avium_104 PFFAI-AKEINVQFVLAYTPE----EFGDSLRALAEGDIDVAPLITGEVG
MSMEG_6108|M.smegmatis_MC2_155 PLYAA-VKQISVSFVFGYDPA----EFASSLRAIAEGDIDVRPMITGSVP
MAB_0526c|M.abscessus_ATCC_199 PYFGI-AKEVAVQFVLAYEPA----EFSETLRRIAHGEIDVAPLVTGEVG
Mb3753|M.bovis_AF2122/97 PAMAI-NKEINLRFVLGYTPL----EFRDTLHMLADGKVNAAPLITGTVG
Rv3726|M.tuberculosis_H37Rv PAMAI-NKEINLRFVLGYTPL----EFRDTLHMLADGKVNAAPLITGTVG
Mvan_5762|M.vanbaalenii_PYR-1 PAMAI-GKEIDLRFVFGYTPL----EFRDTLHMLADGKLDASALVTGTVG
Mflv_0638|M.gilvum_PYR-GCK INRSV-MREANIGFSFCYEAQR---QVPAILDLLATGAINLGELITDEIP
TH_4631|M.thermoresistible__bu VQTIQ-NRELILTGVFRYANT-----WPAAIALIRTGRVDVDALITGRYP
MUL_4777|M.ulcerans_Agy99 PTSMFGLTHKQVLGHLGYQNV----DIETLAKLVSLGRLDLSRSISEIVP
MLBr_01784|M.leprae_Br4923 MPLLDFFSHGGSLKSSWYGDCLPERDFPTLVDLYLQGRLPLGKFVSERIG
. * * : ::
MMAR_5116|M.marinum_M LVGVGAA-FDDLAD-PERHCKVLVTP-------------
MAV_0537|M.avium_104 LDGVGAA-FDDLAD-PERHCKVLVTP-------------
MSMEG_6108|M.smegmatis_MC2_155 LDGMADA-FDELAT-PDAHCKILVTP-------------
MAB_0526c|M.abscessus_ATCC_199 LEDVGSA-FADLGD-PERHCKILVVP-------------
Mb3753|M.bovis_AF2122/97 LPGVAAA-FDALGD-PEAHAKIMIDPKSNAASPQPFRVE
Rv3726|M.tuberculosis_H37Rv LPGVAAA-FDALGD-PEAHAKIMIDPKSNAASPQPFRVE
Mvan_5762|M.vanbaalenii_PYR-1 LGGVAAA-FDALGD-PETHAKILIDPAGTAVTP------
Mflv_0638|M.gilvum_PYR-GCK LDAVVSQGLEELRVNRDAHVKILIDPSA-----------
TH_4631|M.thermoresistible__bu LEKTAEA-LESDRIPGNIKSVVEVGTS------------
MUL_4777|M.ulcerans_Agy99 LEDLSLG-IEKLERQDGNPIRILVQP-------------
MLBr_01784|M.leprae_Br4923 LGDVEEA-FHKIHGGKVLRSVVML---------------
* : : :