For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MKAVTCTNAKLEVVDRPSPAPAKGQLLLDVLRCGICGSDLHARLHCDELADVMAESGYHAFMRSNQQVVF GHEFCGEVVDYGPGTRRTPRRGTPVVAMPLLRRGNKEVHGIGLSTMAPGAYAERLVVEQSLTFPVPNGLA PEIAALTEPMAVGWHAVRRGEVGKGDVAIVIGCGPIGLAVICMLKSRGVHTVIASDFSPGRRALATACGA DSVVDPVQDSPYAVAAGLGQGNRHLQSILDAFDLAVGTVERLQRLRLPWWHLWRAAEAAGAATPKRPVIF ECVGVPGIIDGIIASAPLFSRVVVVGVCMGSDHIRPAMAINKEINLRFVLGYTPLEFRDTLHMLADGKVN AAPLITGTVGLPGVAAAFDALGDPEAHAKIMIDPKSNAASPQPFRVE
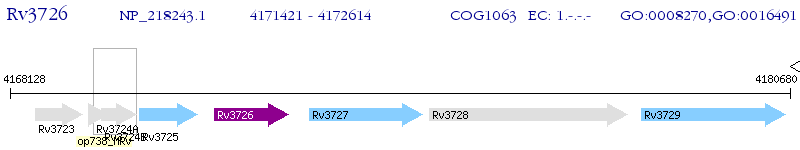
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv3726 | - | - | 100% (397) | POSSIBLE DEHYDROGENASE |
| M. tuberculosis H37Rv | Rv1895 | - | 9e-16 | 27.70% (278) | dehydrogenase |
| M. tuberculosis H37Rv | Rv1530 | adh | 3e-10 | 27.52% (367) | alcohol dehydrogenase adh |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3753 | - | 0.0 | 99.75% (397) | putative dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0638 | - | 2e-26 | 25.96% (389) | alcohol dehydrogenase |
| M. leprae Br4923 | MLBr_01784 | adhE2 | 8e-11 | 25.86% (232) | putative alcohol dehydrogenase (Zn dependent) |
| M. abscessus ATCC 19977 | MAB_0526c | - | 1e-71 | 41.71% (374) | hypothetical protein MAB_0526c |
| M. marinum M | MMAR_3328 | adhE2_1 | 3e-78 | 43.38% (385) | zinc-dependent alcohol dehydrogenase AdhE2_1 |
| M. avium 104 | MAV_0537 | - | 3e-75 | 42.90% (373) | zinc-binding dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6108 | - | 1e-68 | 39.33% (389) | zinc-binding dehydrogenase |
| M. thermoresistible (build 8) | TH_4627 | - | 1e-12 | 26.49% (302) | PUTATIVE Alcohol dehydrogenase, zinc-binding domain protein |
| M. ulcerans Agy99 | MUL_2962 | - | 6e-17 | 28.30% (265) | dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_5762 | - | 1e-142 | 64.45% (391) | alcohol dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Rv3726|M.tuberculosis_H37Rv ----MKAVTCT--NAKLEVVD-RPSPAPAKGQLLLDVLRCGICGSDLHAR
Mb3753|M.bovis_AF2122/97 ----MKAVTCT--NAKLEVVD-RPSPAPAKGQLLLDVLRCGICGSDLHAR
Mvan_5762|M.vanbaalenii_PYR-1 ----MKAVSCV--HGTLEVVD-LPSPRPGEGQLVLDVLRCGICGSDLHAK
MMAR_3328|M.marinum_M ----MRAVLMQ--DSQLQVAE-VPEPVPGPGEVLVDVLACGICGSDLHCA
MAV_0537|M.avium_104 ----MRAAVLR--DGRMVYRDDVPEPIPETGQVLVGVRACGICGSDLHFA
MSMEG_6108|M.smegmatis_MC2_155 ----MRAAVLR--QGRMVVRDDVAEPVPGPGQVLVEVKACGICGSDLHFA
MAB_0526c|M.abscessus_ATCC_199 ----MRASVLR--NGSMVYRDDVPSPTPGEGQVLVKVSACGICGSDLHFA
MLBr_01784|M.leprae_Br4923 MSQTVRGVISRKKDEPVELVD-IVIPDPGPGEAVVDVTACGVCHTDLTYR
TH_4627|M.thermoresistible__bu MSQSVRGVIARKKDEPVEVVD-IVIPDPGPGEVVVKVTACGVCHTDLHYR
MUL_2962|M.ulcerans_Agy99 ----MRTVVIDG-PGSIRVDNRPDPALPGSDGVIVVVSAAGICGSDLHFY
Mflv_0638|M.gilvum_PYR-GCK ----MRAAIYHG--REDVRIEELPDPSPRAGEVVIEVARAGICGTDLHEY
:: : . * . :: * .*:* :**
Rv3726|M.tuberculosis_H37Rv LHC-DELADVMAESG----YHAFMRSNQQVVFGHEFCGEVVDYGPGTRRT
Mb3753|M.bovis_AF2122/97 LHC-DELADVMAESG----YHAFMRSNQQVVFGHEFCGEVVDYGPGTRRT
Mvan_5762|M.vanbaalenii_PYR-1 DHA-DELTEVMAEGG----YHDFVRSDTPVVMGHEFCGVVADRGRKVGKE
MMAR_3328|M.marinum_M AHG-PEFNSATQAAG----GFDMMDLSRPVVFGHEFVGRIAAYGPDTVQR
MAV_0537|M.avium_104 AHG-AQVLQMSDRVAGGAAGMR-VDLDHDIFMGHEFSAEILEAGPDT-ET
MSMEG_6108|M.smegmatis_MC2_155 QHG-EDLLASTAQLEGVANQNDDVDLSRDIFMGHEFSAEILEAGPST-DT
MAB_0526c|M.abscessus_ATCC_199 KHG-ARAIELSKQMVGMPSLTDGVDLSTDVFMGHEFAAEVIEAGPRT-QA
MLBr_01784|M.leprae_Br4923 EGG-INDRYPFLLGHEAAGTVEVVGPGVTAVEPGDFVILNWRAVCGQCRA
TH_4627|M.thermoresistible__bu EGG-INDEFPFLLGHEAAGTVESVGSGVTNVEPGDFVVLNWRAVCGQCRA
MUL_2962|M.ulcerans_Agy99 EGE-YPLAEPVALGHEAVGTVVEKGPDVHTVEVGDQVMVSSVAGCGACAG
Mflv_0638|M.gilvum_PYR-GCK IAGPMHAAPGVVIGHEYSGTVVGVGSGVREFTEGDRVCGVGVFGCGECGF
. . :
Rv3726|M.tuberculosis_H37Rv PRRGTPVVAMPLLRRGNKEVHG---IGLSTMAPGAYAERLVVEQSLTFPV
Mb3753|M.bovis_AF2122/97 PRRGTPVVAMPLLRRGNKEVHG---IGLSTMAPGAYAERLVVEQSLTFPV
Mvan_5762|M.vanbaalenii_PYR-1 FKPGSTVVAFPLMR-AHGGVHL---TGLSPLAPGGYAEQVVAEAALTFVV
MMAR_3328|M.marinum_M IPVGRRVVSAPFLLREQKVTLG---FAG-PEAPGGYSERMLLSEPLLIEV
MAV_0537|M.avium_104 HPPGTLVTSLPVLLSAKGVEP----IVYSNTTIGGYAERMLLSAPLLLPI
MSMEG_6108|M.smegmatis_MC2_155 YAPGTLVTSVPVLISPQGMDM----IVYSNTTLGGYAERMLLSAPLLLPV
MAB_0526c|M.abscessus_ATCC_199 PASGTAVTSIPILLSAKGLEP----IVYSNTTLGGYAEQMLLSAPLLLPI
MLBr_01784|M.leprae_Br4923 CKRGRPSYCFDTFNAQQKMTLIDGTELTPALGIGAFADKTLVHSGQCTKV
TH_4627|M.thermoresistible__bu CKRGRPHLCFDTFNATQKMTLGDGTELTPALGIGAFAEKTLVHEGQCTKV
MUL_2962|M.ulcerans_Agy99 CATRDPAMCVSGPQIFGSGALG---------GAQADLLAVPATDFQVLKI
Mflv_0638|M.gilvum_PYR-GCK CKQGAEALCGAVGFIGFA--------VN-----GALARYASLPTKALFRI
. . :
Rv3726|M.tuberculosis_H37Rv PNGLAPEIAALTEP--MAVGWHAVRRGEVGKGDVAIVIGCGPIGLAVICM
Mb3753|M.bovis_AF2122/97 PNGLAPEIAALTEP--MAVGWHAVRRGEVGKGDVAIVIGCGPIGLAVICM
Mvan_5762|M.vanbaalenii_PYR-1 PNGLDPATAALTEP--MAVAYHAVRRSDVTTKDVAIVLGCGPVGLAVICH
MMAR_3328|M.marinum_M PDHVPTEVAALTEP--LAVAYRTVARVDTTTDDVALVVGCGPIGLAVIAV
MAV_0537|M.avium_104 PNGLDVKHAALTEP--MAVGLHAVNKSNITPAETALVLGCGPIGIAIIAA
MSMEG_6108|M.smegmatis_MC2_155 PNGLDAGPAALTEP--MAVGLHAVNTSRIERNETALVIGCGPVGIAVIAA
MAB_0526c|M.abscessus_ATCC_199 PNGLDPRHAALTEP--MAVGLHAVNKSAIQPGTGAIVIGCGPVGIAVIAA
MLBr_01784|M.leprae_Br4923 DPAADPAVAGLLGCGVMAGLGAAINTAAVSRDDTVAVIGCGGVGDAAISG
TH_4627|M.thermoresistible__bu DPAADPAVAGLLGCGVMAGVGAAINTGAVSRDDTVAVIGCGGVGDAAIAG
MUL_2962|M.ulcerans_Agy99 PAAITTEQALLLTDN-LATGWAAARRADIPFGATVAVIGLGAVGLCAVRS
Mflv_0638|M.gilvum_PYR-GCK PDEISLAEAAVVEP--IASAYHAVRRSGLAAGGTVFIAGAGPIGLALVQF
* : :* : . : * * :* . :
Rv3726|M.tuberculosis_H37Rv LKSRGVHTVIASDFSPGRRALATACGADSVVDPVQDSPYAVAAGLGQGNR
Mb3753|M.bovis_AF2122/97 LKSRGVHTVIASDFSPGRRALATACGADSVVDPVQDSPYAVAAGLGQGNR
Mvan_5762|M.vanbaalenii_PYR-1 LKALGVRTIVASDFSPGRRALAARCGADVVVDPAVDSPYEAVPAKQRG--
MMAR_3328|M.marinum_M LKMKGVGPIVASDFSPERRALATQLGADVVLDPNENSPFEAF--------
MAV_0537|M.avium_104 LATRGVESIVASDFSPKRRQLATAMGAHRTLDPAQGSPFDSVK-------
MSMEG_6108|M.smegmatis_MC2_155 LKLLGVEDIVASDFSPTRRRLAAAMGAHVTLDPAQDSPFHQCR-------
MAB_0526c|M.abscessus_ATCC_199 LHNLGIEPIVASDYSSARRDLAQRMGAHQVVDPKIEPTFHAWSRIGAG--
MLBr_01784|M.leprae_Br4923 AALVGANRIIAVDIDDTKLEWARTFGATHTVNALELEVVKTIQDLTSG--
TH_4627|M.thermoresistible__bu AALVGAKKIIAVDTDAKKLDWAREFGATHTIDASSTDVVEAIQELTDG--
MUL_2962|M.ulcerans_Agy99 ALAQGAATVFAVDRVQGRLQRAAEWGATAVASPAAEAIVAATSG------
Mflv_0638|M.gilvum_PYR-GCK SLAQGATQVIVSEVSATRRVAAHRVGATRVIDPLAEDAVEVVRTLTNG--
* :.. : : * ** . ..
Rv3726|M.tuberculosis_H37Rv HLQSILDAFDLAVGTVERLQRLRLPWWHLWRAAEAAGAATPKRP-VIFEC
Mb3753|M.bovis_AF2122/97 HLQSILDAFDLAVGTVESLQRLRLPWWHLWRAAEAAGAATPKRP-VIFEC
Mvan_5762|M.vanbaalenii_PYR-1 -LTELPALYELGVGSMEKLRKIPG-WSHVYRVADRLGGTAPKRP-VIFEC
MMAR_3328|M.marinum_M ----------MAAAVTDDPARMAP--------AHTVFGQLPLRPTVAFEC
MAV_0537|M.avium_104 -------------------------------------------PAVVFEA
MSMEG_6108|M.smegmatis_MC2_155 -------------------------------------------PAVVFEA
MAB_0526c|M.abscessus_ATCC_199 -------------------------------------------APVIFEA
MLBr_01784|M.leprae_Br4923 -----------------------------------------FGVDVVIDT
TH_4627|M.thermoresistible__bu -----------------------------------------FGADVVVDA
MUL_2962|M.ulcerans_Agy99 -----------------------------------------RGADSVIDA
Mflv_0638|M.gilvum_PYR-GCK -----------------------------------------NGVDISFDA
.:
Rv3726|M.tuberculosis_H37Rv VGVPGIIDGIIASAPLFSRVVVVGVCMGSDHIRP----AMAINKEINLRF
Mb3753|M.bovis_AF2122/97 VGVPGIIDGIIASAPLFSRVVVVGVCMGSDHIRP----AMAINKEINLRF
Mvan_5762|M.vanbaalenii_PYR-1 VGVPGMIDGIIGAAPLASRVVVVGVCMGDDKLRP----AMAIGKEIDLRF
MMAR_3328|M.marinum_M VGVPGLIQQLIAGVPPGTRIGVAGINMSSDHFEP----AQCIAKELDIRF
MAV_0537|M.avium_104 VGVPGIIDDVLLRARPGTRLVVAGVCMQPDTVHP----FFAIAKEINVQF
MSMEG_6108|M.smegmatis_MC2_155 VGAPGIIDDILRRCPAGTRVVVAGVCMQPDTVHP----LYAAVKQISVSF
MAB_0526c|M.abscessus_ATCC_199 VGVPGILNEVLRDAPRSARVVVVGVCMEPDAITP----YFGIAKEVAVQF
MLBr_01784|M.leprae_Br4923 VGRPETWKQAFYARDLAGTVVLVGVPTPDMRLDMPLLDFFSHGGSLKSSW
TH_4627|M.thermoresistible__bu VGRPETWEQAFYARDLAGTVVLVGVPTPDMRIDMPLIDFFSRGGSLKSSW
MUL_2962|M.ulcerans_Agy99 VDTDASMTDALNAVRAGGTVSVVGVHDLQPFPLP---ALTCLLRSITLRL
Mflv_0638|M.gilvum_PYR-GCK AGVQPALDAALGVLRPRGRLMVVAIWEAPAGIDIN----RSVMREANIGF
.. : : :..: .
Rv3726|M.tuberculosis_H37Rv VLGYTP-LEFRDTLHMLADGKVNAAPLITGTVGLPGVAAA--FDALGDPE
Mb3753|M.bovis_AF2122/97 VLGYTP-LEFRDTLHMLADGKVNAAPLITGTVGLPGVAAA--FDALGDPE
Mvan_5762|M.vanbaalenii_PYR-1 VFGYTP-LEFRDTLHMLADGKLDASALVTGTVGLGGVAAA--FDALGDPE
MMAR_3328|M.marinum_M SLYYTL-EEFAQTFAHLAAGDFDVAPLLTGTVALEGVADA--FERLRNNP
MAV_0537|M.avium_104 VLAYTP-EEFGDSLRALAEGDIDVAPLITGEVGLDGVGAA--FDDLADPE
MSMEG_6108|M.smegmatis_MC2_155 VFGYDP-AEFASSLRAIAEGDIDVRPMITGSVPLDGMADA--FDELATPD
MAB_0526c|M.abscessus_ATCC_199 VLAYEP-AEFSETLRRIAHGEIDVAPLVTGEVGLEDVGSA--FADLGDPE
MLBr_01784|M.leprae_Br4923 YGDCLPERDFPTLVDLYLQGRLPLGKFVSERIGLGDVEEA--FHKIHGGK
TH_4627|M.thermoresistible__bu YGDCLPERDFPTLINLYLQGRLPLEKFVTERIALDEVEGA--FEKMQRGE
MUL_2962|M.ulcerans_Agy99 TIAPVQ-RTWSELIPLLESGRLDVDGIFTTSLPLDEAAKG--YSIAGSRS
Mflv_0638|M.gilvum_PYR-GCK SFCYEAQRQVPAILDLLATGAINLGELITDEIPLDAVVSQGLEELRVNRD
. * . :.: : *
Rv3726|M.tuberculosis_H37Rv AHAKIMIDPKSNAASPQPFRVE
Mb3753|M.bovis_AF2122/97 AHAKIMIDPKSNAASPQPFRVE
Mvan_5762|M.vanbaalenii_PYR-1 THAKILIDPAGTAVTP------
MMAR_3328|M.marinum_M HDAKILLDPTR-----------
MAV_0537|M.avium_104 RHCKVLVTP-------------
MSMEG_6108|M.smegmatis_MC2_155 AHCKILVTP-------------
MAB_0526c|M.abscessus_ATCC_199 RHCKILVVP-------------
MLBr_01784|M.leprae_Br4923 VLRSVVML--------------
TH_4627|M.thermoresistible__bu VLRSVVVLK-------------
MUL_2962|M.ulcerans_Agy99 GNDVKVLLKV------------
Mflv_0638|M.gilvum_PYR-GCK AHVKILIDPSA-----------
::