For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RTVVIDGPGSIRVDNRPDPALPGSDGVIVVVSAAGICGSDLHFYEGEYPLAEPVALGHEAVGTVVEKGPD VHTVEVGDQVMVSSVAGCGACAGCATRDPAMCVSGPQIFGSGALGGAQADLLAVPATDFQVLKIPAAITT EQALLLTDNLATGWAAARRADIPFGATVAVIGLGAVGLCAVRSALAQGAATVFAVDRVQGRLQRAAEWGA TAVASPAAEAIVAATSGRGADSVIDAVDTDASMTDALNAVRAGGTVSVVGVHDLQPFPLPALTCLLRSIT LRLTIAPVQRTWSELIPLLESGRLDVDGIFTTSLPLDEAAKGYSIAGSRSGNDVKVLLKV*
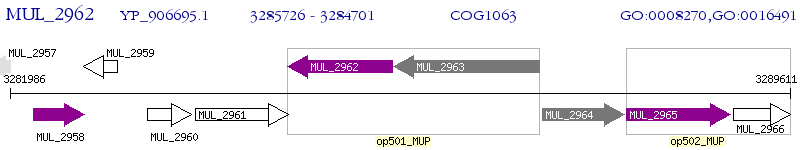
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_2962 | - | - | 100% (341) | dehydrogenase |
| M. ulcerans Agy99 | MUL_4777 | - | 2e-29 | 29.94% (324) | zinc-dependent alcohol dehydrogenase |
| M. ulcerans Agy99 | MUL_0470 | adhB | 4e-27 | 30.63% (333) | zinc-containing alcohol dehydrogenase NAD- dependent AdhB |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1929 | - | 1e-107 | 84.42% (231) | dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4517 | - | 1e-131 | 69.32% (339) | alcohol dehydrogenase |
| M. tuberculosis H37Rv | Rv1895 | - | 1e-158 | 80.59% (340) | dehydrogenase |
| M. leprae Br4923 | MLBr_01784 | adhE2 | 2e-26 | 36.82% (258) | putative alcohol dehydrogenase (Zn dependent) |
| M. abscessus ATCC 19977 | MAB_0983c | - | 5e-32 | 32.19% (320) | zinc-containing alcohol dehydrogenase |
| M. marinum M | MMAR_2790 | - | 0.0 | 98.24% (341) | dehydrogenase |
| M. avium 104 | MAV_2804 | - | 1e-157 | 80.00% (340) | zinc-binding dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6616 | - | 7e-50 | 34.52% (394) | S-(hydroxymethyl)glutathione dehydrogenase |
| M. thermoresistible (build 8) | TH_4349 | - | 6e-34 | 35.69% (325) | PUTATIVE Alcohol dehydrogenase GroES domain protein |
| M. vanbaalenii PYR-1 | Mvan_1841 | - | 1e-132 | 70.21% (339) | alcohol dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4517|M.gilvum_PYR-GCK ----------MRTVVVDSQRSIRVDTRPDPRLPGPDGVIVDVTASAICGS
Mvan_1841|M.vanbaalenii_PYR-1 ----------MRAVVIDSERRIRVETRPDPQLPGPDGAIVEVTATAICGS
MUL_2962|M.ulcerans_Agy99 ----------MRTVVIDGPGSIRVDNRPDPALPGSDGVIVVVSAAGICGS
MMAR_2790|M.marinum_M ----------MRTVVIDGPGSIRVDNRPDPALPGSDGVIVAVSAAGICGS
Mb1929|M.bovis_AF2122/97 --------------------------------------------------
Rv1895|M.tuberculosis_H37Rv ----------MRAVVIDGAGSVRVNTQPDPALPGPDGVVVAVTAAGICGS
MAV_2804|M.avium_104 ----------MRTVVVDGPRSIRVDTRPDPVLPGPDGAIVEVSAAGICGS
MSMEG_6616|M.smegmatis_MC2_155 ----------MKAVTWHGRRDVRVDNVPDPKIEQPTDAIIEVTSTNICGS
TH_4349|M.thermoresistible__bu ----------MKALRLMDWKSEPELVEVPDPTPGPGQVLVKIGGAGACHS
MLBr_01784|M.leprae_Br4923 ------MSQTVRGVISRKKDEPVELVDIVIPDPGPGEAVVDVTACGVCHT
MAB_0983c|M.abscessus_ATCC_199 MRIRSAVLHAAPIERPYRDTHPLKVIELELDPPGPGEVLVRIEAAGLCHS
Mflv_4517|M.gilvum_PYR-GCK DLHFLDGH----YPIDQPVPIGHEAVGVVVETGSDVTRFTNGDRVLVSSV
Mvan_1841|M.vanbaalenii_PYR-1 DLHFLEGH----YPIDQPVSIGHEAVGVVVETGTTVAGFKVGDRVLVSSV
MUL_2962|M.ulcerans_Agy99 DLHFYEGE----YPLAEPVALGHEAVGTVVEKGPDVHTVEVGDQVMVSSV
MMAR_2790|M.marinum_M DLHFYEGE----YPLAEPVALGHEAVGTVVEKGPDVHTVEVGDQVMVSSV
Mb1929|M.bovis_AF2122/97 --------------------------------------------------
Rv1895|M.tuberculosis_H37Rv DLHFYEGE----YPFTEPVALGHEAVGTIVEAGPQVRTVGVGDLVMVSSV
MAV_2804|M.avium_104 DLHFYEAD----FPMPDPIALGHEAIGTVVEAGPQVRNVKVGDQVMVSSV
MSMEG_6616|M.smegmatis_MC2_155 DLHLYEVLG---AFMNEGDILGHEPMGIVREVGSAVDGLSVGDRVVIPFQ
TH_4349|M.thermoresistible__bu DLHLMHDFEPGMMPWQLPFTLGHENAGWVAAVGAGVGTVTEGDAVAVYGP
MLBr_01784|M.leprae_Br4923 DLTYREGG----INDRYPFLLGHEAAGTVEVVGPGVTAVEPGDFVILNWR
MAB_0983c|M.abscessus_ATCC_199 DLSVVDGN----RIRPVPMALGHEAAGVIAAVGPGVRDVSPGDHVVLVYV
Mflv_4517|M.gilvum_PYR-GCK TGCGRCAGCLTLDPVKCVH--------GPQIFG-----------TGLLGG
Mvan_1841|M.vanbaalenii_PYR-1 AGCGRCAGCRTQDPVKCVQ--------GPQIFG-----------SGLLGG
MUL_2962|M.ulcerans_Agy99 AGCGACAGCATRDPAMCVS--------GPQIFG-----------SGALGG
MMAR_2790|M.marinum_M AGCGACAGCATRDPAMCVS--------GPQIFG-----------SGALGG
Mb1929|M.bovis_AF2122/97 -----------------------------MIFG-----------AGVLGG
Rv1895|M.tuberculosis_H37Rv AGCGVCPGCETHDPVMCFS--------GPMIFG-----------AGVLGG
MAV_2804|M.avium_104 AGCGSCAGCATRDPVLCHS--------GFQIFG-----------GGVLGG
MSMEG_6616|M.smegmatis_MC2_155 ISCGHCFMCDQKLYTQCETTQVREQGMGAALFGYSE-------LYGSVPG
TH_4349|M.thermoresistible__bu WGCGTCERCMGGAENYCEN------PAGAPVLSGG--------AGLGMDG
MLBr_01784|M.leprae_Br4923 AVCGQCRACKRGRPSYCFDTFNAQ-QKMTLIDG---------TELTPALG
MAB_0983c|M.abscessus_ATCC_199 PSCGACRYCSSGRPALCPDAAAAN-GRGDLIRGGTRLHDLDGTEVRHHLG
: . *
Mflv_4517|M.gilvum_PYR-GCK AQSDLLAVPAADFQLLAVPDGIGTEEALLLTDNLPTGWAAAKR--ADIPV
Mvan_1841|M.vanbaalenii_PYR-1 AQADLMAVPAADFQLLAIPDGIDTEQALLLTDNLATGWAAAKR--ADIPV
MUL_2962|M.ulcerans_Agy99 AQADLLAVPATDFQVLKIPAAITTEQALLLTDNLATGWAAARR--ADIPF
MMAR_2790|M.marinum_M AQADLLAVPAADFQVLKIPAAITTEQALLLTDNLATGWAAARR--ADILF
Mb1929|M.bovis_AF2122/97 AQADLLAVPAADFQVLKIPEGITTEQALLLTDNLATGWAAAQR--ADISF
Rv1895|M.tuberculosis_H37Rv AQADLLAVPAADFQVLKIPEGITTEQALLLTDNLATGWAAAQR--ADISF
MAV_2804|M.avium_104 AQADLLAVPAADFQLLKIPEGISTEQALLLTDNLATGWAAAQR--ADIPF
MSMEG_6616|M.smegmatis_MC2_155 GQAQYLRVPQAQFTHIKVPDGPPDSRYVYLSDVLPTAWQSVAY--AEIPK
TH_4349|M.thermoresistible__bu GMAEYLLVP-AERLVVKLPESMTPVTAAALTDAGLTPYHAIRRSWPKLTP
MLBr_01784|M.leprae_Br4923 IGAFADKTLVHSGQCTKVDPAADPAVAGLLGCGVMAGLGAAIN-TAAVSR
MAB_0983c|M.abscessus_ATCC_199 VSGFADHAVVDRNSVVVIDENVPLDTAALFGCAMLTGFGAVTH-TASVRP
. . : : : : . :
Mflv_4517|M.gilvum_PYR-GCK GGTVAVIGAGAVGQCALRSALALGAARVFAVDPVVARRERAAAAGATPMD
Mvan_1841|M.vanbaalenii_PYR-1 GGTVAVIGAGAVGQCALRSAYALGAATVFAVDPVAARRDRAAAAGARAVP
MUL_2962|M.ulcerans_Agy99 GATVAVIGLGAVGLCAVRSALAQGAATVFAVDRVQGRLQRAAEWGATAVA
MMAR_2790|M.marinum_M GATVAVIGLGAVGLCAVRSALAQGAATVFAVDRVQGRLQRASEWGATAVA
Mb1929|M.bovis_AF2122/97 GSAVAVIGLGAVGLCALRSAFIHGAATVFAVDRVKGRLQRAATWGATPIP
Rv1895|M.tuberculosis_H37Rv GSAVAVIGLGAVGLCALRSAFIHGAATVFAVDRVKGRLQRAATWGATPIP
MAV_2804|M.avium_104 GGTVAVIGLGAVGLCALRSALFQGAATVFAVDQVDGRLARAARWGATPLK
MSMEG_6616|M.smegmatis_MC2_155 GGTVTVLGLGPIGDMAARIAHHLG-YEVFAVDRIPERLRRAADRGIQAID
TH_4349|M.thermoresistible__bu TSTAVVIGVGGLGHVAVQLVKATTAARLIAVDNRSEALELAREMGADHVV
MLBr_01784|M.leprae_Br4923 DDTVAVIGCGGVGDAAISGAALVGANRIIAVDIDDTKLEWARTFGATHTV
MAB_0983c|M.abscessus_ATCC_199 GDSVAVLGLGGVGLAVVMSAAAAGAGEVLAIDPVPAKRELALELGATSAC
:..*:* * :* . . ::*:* * *
Mflv_4517|M.gilvum_PYR-GCK S-----PAAQTILEATDGLGADSVIDAVGTDAS-----------------
Mvan_1841|M.vanbaalenii_PYR-1 A-----PAAAAILEATGGLGVDSVIDAVGTDTS-----------------
MUL_2962|M.ulcerans_Agy99 S-----PAAEAIVAATSGRGADSVIDAVDTDAS-----------------
MMAR_2790|M.marinum_M S-----PAAEAIVAATGGRGADSVIDAVGTDAS-----------------
Mb1929|M.bovis_AF2122/97 S-----PAAETILAATRGRGADSVIDAVGTDAS-----------------
Rv1895|M.tuberculosis_H37Rv S-----PAAETILAATRGRGADSVIDAVGTDAS-----------------
MAV_2804|M.avium_104 A-----PALEAILAATGGRGADAVIDAVATDAS-----------------
MSMEG_6616|M.smegmatis_MC2_155 LGAIDEPLGDVIRSMTHGRGTDAVIDAVGMEAHGSPVAKIMQTAAAALPD
TH_4349|M.thermoresistible__bu RS--DEQAVEAIWELTGGRGADVLLDFVGADAT-----------------
MLBr_01784|M.leprae_Br4923 NA-LELEVVKTIQDLTSGFGVDVVIDTVGRPET-----------------
MAB_0983c|M.abscessus_ATCC_199 AP-DEAPG-----------GHDWVFEVVGSAAV-----------------
* * ::: *
Mflv_4517|M.gilvum_PYR-GCK -----------------IDDALAGVRTGGTVSVVGVHDLTP-YPLPALVC
Mvan_1841|M.vanbaalenii_PYR-1 -----------------LDDALACVRTGGTVSIVGVHDLQP-YPLPALVC
MUL_2962|M.ulcerans_Agy99 -----------------MTDALNAVRAGGTVSVVGVHDLQP-FPLPALTC
MMAR_2790|M.marinum_M -----------------MTDALNAVRAGGTVSVVGVHDLQP-FPLPALTC
Mb1929|M.bovis_AF2122/97 -----------------MSDALNAVRPGGTVSVVGVHDLQP-FPLPALTC
Rv1895|M.tuberculosis_H37Rv -----------------MSDALNAVRPGGTVSVVGVHDLQP-FPVPALTC
MAV_2804|M.avium_104 -----------------LTEALNAVRPGGTVSVVGVHDMNP-FPLNALGC
MSMEG_6616|M.smegmatis_MC2_155 AVAKPMMENAGVDRLDALYSAIDTVRRGGTISLIGVYGGTA-DPIPMLTL
TH_4349|M.thermoresistible__bu -----------------IELARAAARPLGDVTIVGIAGGS-----VPLSF
MLBr_01784|M.leprae_Br4923 -----------------WKQAFYARDLAGTVVLVGVPTPDMRLDMPLLDF
MAB_0983c|M.abscessus_ATCC_199 -----------------LEQAYALTGRGGGTVSVGLPHPDARISLPALNI
* * :*: *
Mflv_4517|M.gilvum_PYR-GCK LLRSLTLR---MTTAPVQQTWAELVPLLQAGR-LSVDGIFTDAMPLAEAE
Mvan_1841|M.vanbaalenii_PYR-1 LLRSLTIR---LTTAPVQQTWPELIPLLQAGR-LSVDGIFTGALPLDDAE
MUL_2962|M.ulcerans_Agy99 LLRSITLR---LTIAPVQRTWSELIPLLESGR-LDVDGIFTTSLPLDEAA
MMAR_2790|M.marinum_M LLRSITLR---LTIAPVQRTWSELIPLLESGR-LDVDGIFTTSLPLDEAA
Mb1929|M.bovis_AF2122/97 LLRSITLR---MTMAPVQRTWPELIPLLQSGR-LDVDGIFTTTLPLDEAA
Rv1895|M.tuberculosis_H37Rv LLRSITLR---MTMAPVQRTWPELIPLLQSGR-LDVDGIFTTTLPLDEAA
MAV_2804|M.avium_104 LIRSITLR---MTTAPVQRTWPELIPLLQSGR-LDVDGIFTTTMPLDEAA
MSMEG_6616|M.smegmatis_MC2_155 FDKQIQLR---MGQANVKRWVDDIMPLLGDDDPLGVDDFATHILPIDEAP
TH_4349|M.thermoresistible__bu FSQPYEVS---IQTTYWG-TRPELVELLDLAA-RGLVHVESTTYPLDDAL
MLBr_01784|M.leprae_Br4923 FSHGGSLKSSWYGDCLPERDFPTLVDLYLQGR-LPLGKFVSERIGLGDVE
MAB_0983c|M.abscessus_ATCC_199 VAEGRSILGSYMGSAQPQQDIPAMLALWRAGR-LPVEKLKSDDLPLSDIN
. . : :: * : . : : :
Mflv_4517|M.gilvum_PYR-GCK AAYAAAFSRSGEHLKVHLVP------------------------------
Mvan_1841|M.vanbaalenii_PYR-1 RAYAAAFSRSAEHLKVQLIP------------------------------
MUL_2962|M.ulcerans_Agy99 KGYSIAGSRSGNDVKVLLKV------------------------------
MMAR_2790|M.marinum_M KGYSIAGSRSGNDVKVLLKV------------------------------
Mb1929|M.bovis_AF2122/97 KGYATARARSGEELKVLLTP------------------------------
Rv1895|M.tuberculosis_H37Rv KGYATARARSGEELRFCLRPDSRDVLGAHETVDLYVHVRRCQSVADLQLE
MAV_2804|M.avium_104 IGYATAGSRSGDDVKILLKP------------------------------
MSMEG_6616|M.smegmatis_MC2_155 HAYEIFQKKQDGAVKIILKP------------------------------
TH_4349|M.thermoresistible__bu QAYRDLRAGRVRGRAVIVP-------------------------------
MLBr_01784|M.leprae_Br4923 EAFHKIHGGKVLRSVVML--------------------------------
MAB_0983c|M.abscessus_ATCC_199 IALDALAEGQAVRQILRPGVSA----------------------------
. .
Mflv_4517|M.gilvum_PYR-GCK -------------
Mvan_1841|M.vanbaalenii_PYR-1 -------------
MUL_2962|M.ulcerans_Agy99 -------------
MMAR_2790|M.marinum_M -------------
Mb1929|M.bovis_AF2122/97 -------------
Rv1895|M.tuberculosis_H37Rv GAADGVDGPSMLN
MAV_2804|M.avium_104 -------------
MSMEG_6616|M.smegmatis_MC2_155 -------------
TH_4349|M.thermoresistible__bu -------------
MLBr_01784|M.leprae_Br4923 -------------
MAB_0983c|M.abscessus_ATCC_199 -------------