For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSEHWTVLVTGASSGIGASAARIFAERGHRVFGTSRNPDTITDRVPGVQYLRLDQTDKASIAECVAQAGE VDILVNNAGESQIGALEDVSMDDFEKLYMTNVFGPVALTKAVLPGMRERRRGAVVMVGSMAGTLHVPFRS TYSSAKSALHTVADCLRYEVAPFGITVSTVEPGVIATGIETRRNRIVPDGSPYRDAFRTVDAAMAAREEH GTATDELGRLVVDVALSPSPKPLYAKGSNAPIIMTAQRLLPARLFLKFLARMHGLKVR
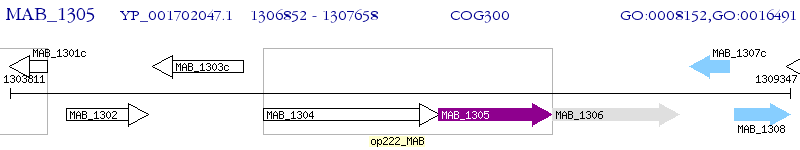
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1305 | - | - | 100% (268) | short-chain dehydrogenase/reductase |
| M. abscessus ATCC 19977 | MAB_3506c | - | 6e-25 | 35.20% (179) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0090c | - | 6e-22 | 34.24% (184) | oxidoreductase EphD |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3199 | - | 2e-22 | 33.64% (217) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_5243 | - | 2e-30 | 35.89% (248) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv3174 | - | 8e-23 | 34.10% (217) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_00862 | ephD | 2e-18 | 36.22% (185) | short chain dehydrogenase |
| M. marinum M | MMAR_3561 | - | 6e-33 | 32.44% (262) | oxidoreductase |
| M. avium 104 | MAV_2098 | - | 3e-37 | 35.53% (273) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_0524 | - | 9e-29 | 39.13% (207) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_3605 | - | 1e-28 | 34.60% (263) | PUTATIVE NAD dependent epimerase/dehydratase family protein |
| M. ulcerans Agy99 | MUL_2794 | - | 2e-33 | 32.44% (262) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_1012 | - | 4e-30 | 35.48% (248) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_1305|M.abscessus_ATCC_1997 --------------------------------------------------
TH_3605|M.thermoresistible__bu --------------------------------------------------
Mflv_5243|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1012|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_3561|M.marinum_M --------------------------------------------------
MUL_2794|M.ulcerans_Agy99 --------------------------------------------------
MAV_2098|M.avium_104 --------------------------------------------------
MSMEG_0524|M.smegmatis_MC2_155 --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00862|M.leprae_Br4923 MPATQSIHKVPHQFVESPDGVRLAIYQSGQPEGPAIVLVHGFPDSHVLWD
MAB_1305|M.abscessus_ATCC_1997 --------------------------------------------------
TH_3605|M.thermoresistible__bu --------------------------------------------------
Mflv_5243|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1012|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_3561|M.marinum_M --------------------------------------------------
MUL_2794|M.ulcerans_Agy99 --------------------------------------------------
MAV_2098|M.avium_104 --------------------------------------------------
MSMEG_0524|M.smegmatis_MC2_155 --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00862|M.leprae_Br4923 GVVPLLAKRFRIVRYDNRGVGQSSVPKPVSAYTMTRFADDFAAVIDELSP
MAB_1305|M.abscessus_ATCC_1997 --------------------------------------------------
TH_3605|M.thermoresistible__bu --------------------------------------------------
Mflv_5243|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1012|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_3561|M.marinum_M --------------------------------------------------
MUL_2794|M.ulcerans_Agy99 --------------------------------------------------
MAV_2098|M.avium_104 --------------------------------------------------
MSMEG_0524|M.smegmatis_MC2_155 --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DRPVHVLAHDWGSVGVWEYLSRSGASDRVASFTSVSGPSQDQLVDYILSG
MAB_1305|M.abscessus_ATCC_1997 --------------------------------------------------
TH_3605|M.thermoresistible__bu --------------------------------------------------
Mflv_5243|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1012|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_3561|M.marinum_M --------------------------------------------------
MUL_2794|M.ulcerans_Agy99 --------------------------------------------------
MAV_2098|M.avium_104 --------------------------------------------------
MSMEG_0524|M.smegmatis_MC2_155 --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LRRPWRPRTFARAVSQLFRLTYMVLFSIPVLVPMVLWIALSSATIRRTMV
MAB_1305|M.abscessus_ATCC_1997 --------------------------------------------------
TH_3605|M.thermoresistible__bu --------------------------------------------------
Mflv_5243|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1012|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_3561|M.marinum_M --------------------------------------------------
MUL_2794|M.ulcerans_Agy99 --------------------------------------------------
MAV_2098|M.avium_104 --------------------------------------------------
MSMEG_0524|M.smegmatis_MC2_155 --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DNISTDQIHHSATLARDAAHSVKTYPANYFRAFSGRRRGKGVPVVNVPVQ
MAB_1305|M.abscessus_ATCC_1997 --------------------------------------------------
TH_3605|M.thermoresistible__bu --------------------------------------------------
Mflv_5243|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1012|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_3561|M.marinum_M --------------------------------------------------
MUL_2794|M.ulcerans_Agy99 --------------------------------------------------
MAV_2098|M.avium_104 --------------------------------------------------
MSMEG_0524|M.smegmatis_MC2_155 --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LIVNTMDRYVRPYGYDATARWVPRLWRRDIKAGHFSPMSHPQVMAAAVHD
MAB_1305|M.abscessus_ATCC_1997 ------------------------MSEHWTVLVTGASSGIG-ASAARIFA
TH_3605|M.thermoresistible__bu -------------------VTDRQPAGPRSVVITGASRGLG-LASATHLY
Mflv_5243|M.gilvum_PYR-GCK -----------------------MAT----VLITGCSSGYG-RATALKFL
Mvan_1012|M.vanbaalenii_PYR-1 -----------------------MTT----VLITGCSSGYG-LATAHRFL
MMAR_3561|M.marinum_M -----------------------MTSTPPVALITGVSSGIG-AAIAVRLA
MUL_2794|M.ulcerans_Agy99 -----------------------MTSTPPVALITGVSSGIG-AAIAVRLA
MAV_2098|M.avium_104 -----------------------MTHRSPVVLVTGASSGIG-RAVAKAFA
MSMEG_0524|M.smegmatis_MC2_155 ---------------------------MTVWFITGASRGLG-AQIARTAL
Mb3199|M.bovis_AF2122/97 ----------------------MTSLAERTALVTGANRGMGREYVAQLLG
Rv3174|M.tuberculosis_H37Rv ----------------------MTSLAERTVLVTGANRGMGREYVAQLLG
MLBr_00862|M.leprae_Br4923 LADLVDGKQPSRALLRAQVGRSRDTFGDTLVSVTGAGSGIG-RETALALA
:** . * * *
MAB_1305|M.abscessus_ATCC_1997 ERGHRVFGTSRNPDT----------ITDRVPGVQYLRLDQTDKASIAECV
TH_3605|M.thermoresistible__bu RRGWRVVAAMRSVDAGMARLAETTGAAADDPRLIPVALDLTDAASITAAA
Mflv_5243|M.gilvum_PYR-GCK AEGWNVVATMRNPRPD---------VLPASDRLRVSALDVTDPESIASAV
Mvan_1012|M.vanbaalenii_PYR-1 DEGWNVVATMRTPRQD---------VLPPSDRLRVIALDVTRPDSIAAAV
MMAR_3561|M.marinum_M SAGFRVVGTSRAPQR-----------LAPIPGVETLALDVTDDTAVRSVV
MUL_2794|M.ulcerans_Agy99 SAGFRVVGTSRAPQR-----------LAPIPGVETLALDVTDDTAVRSVV
MAV_2098|M.avium_104 AKGFEVFGTTRNPRT-----------AEPIPGVELVELDVTDAASVDAAA
MSMEG_0524|M.smegmatis_MC2_155 DHGADVAVGVRNPDLL---------PSDIAERAFAVRLDVTHDADISAAV
Mb3199|M.bovis_AF2122/97 RKVAKVYAATRNPLAID----------VSDPRVIPLQLDVTDAVSVAEAA
Rv3174|M.tuberculosis_H37Rv RKVAKVYAATRNPLAID----------VSDPRVIPLQLDVTDAVSVAEAA
MLBr_00862|M.leprae_Br4923 RRGAEVVVSDIDEAAAKN---TAAQIVASGGVAYAYALDVSDAAAVEAFA
* ** : : .
MAB_1305|M.abscessus_ATCC_1997 AQAG----EVDILVNNAGESQIGALEDVSMDDFEKLYMTNVFGPVALTKA
TH_3605|M.thermoresistible__bu KEIEGSVGAPYAVVHNAGISAAGMVEETPLSLWEQMFATTVFGPVALTRE
Mflv_5243|M.gilvum_PYR-GCK ----EAAGPLDAVVNNAGIGAVGAFEATPMSTIRELFDTNTFGVMAMTQA
Mvan_1012|M.vanbaalenii_PYR-1 ----AAAGPIDVLVNNAGIGAVGAFEATPMQTVRELFDTNTFGVMAMTQA
MMAR_3561|M.marinum_M SEVIDRTGRIDVLVNNAGLGIAGAAEESSIDQARSLFDTNFFGLIRLTNE
MUL_2794|M.ulcerans_Agy99 SEVIDRTGHIDVLVNNAGLGIAGAAEESSIDQARSLFDTNFFGLIRLTNE
MAV_2098|M.avium_104 KSVIERTGRIDILVNNAGSGILGAAEERSVAQAQRLIDTNFLGLVRLTNA
MSMEG_0524|M.smegmatis_MC2_155 AAVVERFGRIDVLVNNAGRGLLGALEEVTDAEARSLFDLNVFGLINVTRA
Mb3199|M.bovis_AF2122/97 ----DLATDVGILINNAGISRASSVLDKDTSALRGELETNLFGPLALASA
Rv3174|M.tuberculosis_H37Rv ----DLATDVGILINNAGISRASSVLDKDTSALRGELETNLFGPLALASA
MLBr_00862|M.leprae_Br4923 EQVSAKHGVPDIVVNNAGIGQAGRFLDTPPEEFDRVLAVNLGGVVNCCRA
:::*** . . . * :
MAB_1305|M.abscessus_ATCC_1997 VLPGMRERR-RGAVVMVGSMAGTLHVPFRSTYSSAKSALHTVADCLRYEV
TH_3605|M.thermoresistible__bu LLPGMRRAG-RGRIVLVSSQQGVWGMPSTAPYSAAKGALERWAEAMAGEV
Mflv_5243|M.gilvum_PYR-GCK VVPQMRERR-SGVVVNVTSSTTLAPMPLAAAYTASKSAIEGFTGSLALEL
Mvan_1012|M.vanbaalenii_PYR-1 VVPQMRARR-SGVVVNVTSSVTLAAMPLAAAYTASKAAIEGFTGSLALEL
MMAR_3561|M.marinum_M VLPHMRRRG-SGRIINISSVLGFLPAPYAALYAASKHAVEGYTESLDHEL
MUL_2794|M.ulcerans_Agy99 VLPHMRRRG-SGRIINISSVLGFLPAPYAALYAASKHAVEGYTESLDHEL
MAV_2098|M.avium_104 VLPYLRAQG-GGRVINIGSVLGFLPQPYGALYAGSKHAVEGYSESLDHET
MSMEG_0524|M.smegmatis_MC2_155 VLPHMRASG-GGKLVHIGSRSGFEGEPGVGLYSASKFAVAGISEALSAEL
Mb3199|M.bovis_AF2122/97 FADRIAER--SGAIVNVSSVLAWL--PLGMSYGVSKAAMWSATESMRIEL
Rv3174|M.tuberculosis_H37Rv FADRIAER--SGAIVNVSSVLAWL--PLGMSYGVSKAAMWSATESMRIEL
MLBr_00862|M.leprae_Br4923 FGQRLVERGTGGHIVNVSSMAAYAPLQSLSAYCTSKAATYMFSDCLRAEL
. : * :: : * * :* * : .: *
MAB_1305|M.abscessus_ATCC_1997 APFGITVSTVEPGVIATGIETRRNRIVPDGSP--YRDAFRTVDAAMAARE
TH_3605|M.thermoresistible__bu NPFGIGVTVLVTGVYDTEIITDAGTTDCRDLQGVYARHHRTMDKRGRAAV
Mflv_5243|M.gilvum_PYR-GCK GFFGVRAKLVEPGYGPGTQFTSNGVERMAGLIPAEYQDFAAPVFEEFRSP
Mvan_1012|M.vanbaalenii_PYR-1 GFFGVRAKLVEPGYGPGTQFASNGSHRMADLIPDEYQDFAAPIMESFGSP
MMAR_3561|M.marinum_M REYGVRALLVEPSYTR-TDFESN--MWEADTPQAAYADNRAAVRAVVADK
MUL_2794|M.ulcerans_Agy99 REYGVRALLVEPSYTR-TDFESN--MWEADTPQAAYADNRAAVRAVVANK
MAV_2098|M.avium_104 REFGVRVSVVEPAYTR-TSFEAN--MADADAPIDAYAASREHARRRMAEL
MSMEG_0524|M.smegmatis_MC2_155 APFGIQSMVVEPGVFR-TDFLDGSSLSMPANRIDAYDGTPAHDTLDWVDD
Mb3199|M.bovis_AF2122/97 APRGVQVVGVYVGLVD------TDMGRFADAPKSDPADVVRQVLDGIEAG
Rv3174|M.tuberculosis_H37Rv APRGVQVVGVYVGLVD------TDMGRFADAPKSDPADVVRQVLDGIEAG
MLBr_00862|M.leprae_Br4923 DAVGVGLTTICPGVID------TNIIQSTRIDTPTAKQERVRDRRGQLDM
*: : .
MAB_1305|M.abscessus_ATCC_1997 EHGTATDELGRLVVDVALS-PSPKPLYAKGSNAPIIMTAQRLLPARLFLK
TH_3605|M.thermoresistible__bu AKAARPPEKFALALSKALDNPQPFVKRPAGPSARMLMIANRILPPATLHQ
Mflv_5243|M.gilvum_PYR-GCK G-MVTTPDEVADVVFQAATDTSGRLRFAAGADAVALAQ------------
Mvan_1012|M.vanbaalenii_PYR-1 A-AVTTPDDVAEVVYLAATDASDRLRYAAGADAVALAQ------------
MMAR_3561|M.marinum_M IRNAELPAVVADATYAAATDKRIKLRYPAGRHARQLALLKRIAPAAVLDN
MUL_2794|M.ulcerans_Agy99 IRNAELPAVVADATYAAAIDKRIKLRYPAGRHARQLALLKRIAPAAVLDN
MAV_2098|M.avium_104 MKTADDPAIVAEVVLKAATARKPKLRYPAGPLARRLSLLKKFAPEAVLDR
MSMEG_0524|M.smegmatis_MC2_155 ANHTQLGDPVKGAALIYEVANEDKLPRHLLLGRDAVERWRVKAAQMADDV
Mb3199|M.bovis_AF2122/97 KEDVLADEMSRQVRASLNVPARERIARLMGN-------------------
Rv3174|M.tuberculosis_H37Rv KEDVLADEMSRQVRASLNVPARERIARLMGN-------------------
MLBr_00862|M.leprae_Br4923 MFRLRRYGPDKVADTILSAIKKNKPIRPVAPEAYALYGISRVLPQVLRST
.
MAB_1305|M.abscessus_ATCC_1997 FLARMHGLKVR-----------
TH_3605|M.thermoresistible__bu MIRLSLGLPRFGRLTTREDTHA
Mflv_5243|M.gilvum_PYR-GCK ----------------------
Mvan_1012|M.vanbaalenii_PYR-1 ----------------------
MMAR_3561|M.marinum_M GIRKQTGLNSP-RPTPVLAER-
MUL_2794|M.ulcerans_Agy99 GIRKQTGLNSP-RPTPVLAER-
MAV_2098|M.avium_104 GIRKTNGLPAASRPHSHLQL--
MSMEG_0524|M.smegmatis_MC2_155 TAWEMKSLATAHLA--------
Mb3199|M.bovis_AF2122/97 ----------------------
Rv3174|M.tuberculosis_H37Rv ----------------------
MLBr_00862|M.leprae_Br4923 ARIRVI----------------