For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SKKILITGASSGFGRGAAIELARRGHQVVATAETWPQVRTLRADAAEAGVDLEVIKLNLLDEIDIAHAAG YDPDVLVLNAGVMESGSIIDIPLARVRESFDINVFGHIQLAQGIVPKMVARKSGKVVWTSSMGGILVIPF VGVYCATKHAIEAIAGSMKAELEPHGVKVATVNPGLFGTGFNDTGAESHVQWYDPERAVVPMPSFGDTLA DQFDPQEMIDAMVEIIPADEHLYRTMRPLATIDVAKGWQETEWTQNA*
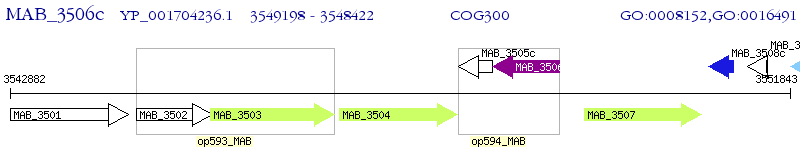
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3506c | - | - | 100% (258) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1305 | - | 5e-25 | 35.20% (179) | short-chain dehydrogenase/reductase |
| M. abscessus ATCC 19977 | MAB_3405 | - | 1e-21 | 33.17% (208) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1896c | - | 2e-17 | 33.69% (187) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_5243 | - | 5e-25 | 38.50% (187) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv1865c | - | 2e-17 | 33.69% (187) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_00862 | ephD | 8e-16 | 27.42% (186) | short chain dehydrogenase |
| M. marinum M | MMAR_3561 | - | 6e-21 | 33.82% (204) | oxidoreductase |
| M. avium 104 | MAV_1976 | - | 2e-25 | 32.64% (242) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_1114 | - | 1e-23 | 31.25% (256) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_4244 | - | 2e-21 | 32.00% (175) | PUTATIVE putative Dehydrogenase/decarboxylase protein |
| M. ulcerans Agy99 | MUL_2794 | - | 1e-21 | 33.82% (204) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_1012 | - | 9e-25 | 38.50% (213) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_1976|M.avium_104 --------------------------------------------------
MSMEG_1114|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_5243|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1012|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_3561|M.marinum_M --------------------------------------------------
MUL_2794|M.ulcerans_Agy99 --------------------------------------------------
Mb1896c|M.bovis_AF2122/97 --------------------------------------------------
Rv1865c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00862|M.leprae_Br4923 MPATQSIHKVPHQFVESPDGVRLAIYQSGQPEGPAIVLVHGFPDSHVLWD
MAB_3506c|M.abscessus_ATCC_199 --------------------------------------------------
TH_4244|M.thermoresistible__bu --------------------------------------------------
MAV_1976|M.avium_104 --------------------------------------------------
MSMEG_1114|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_5243|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1012|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_3561|M.marinum_M --------------------------------------------------
MUL_2794|M.ulcerans_Agy99 --------------------------------------------------
Mb1896c|M.bovis_AF2122/97 --------------------------------------------------
Rv1865c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00862|M.leprae_Br4923 GVVPLLAKRFRIVRYDNRGVGQSSVPKPVSAYTMTRFADDFAAVIDELSP
MAB_3506c|M.abscessus_ATCC_199 --------------------------------------------------
TH_4244|M.thermoresistible__bu --------------------------------------------------
MAV_1976|M.avium_104 --------------------------------------------------
MSMEG_1114|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_5243|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1012|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_3561|M.marinum_M --------------------------------------------------
MUL_2794|M.ulcerans_Agy99 --------------------------------------------------
Mb1896c|M.bovis_AF2122/97 --------------------------------------------------
Rv1865c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DRPVHVLAHDWGSVGVWEYLSRSGASDRVASFTSVSGPSQDQLVDYILSG
MAB_3506c|M.abscessus_ATCC_199 --------------------------------------------------
TH_4244|M.thermoresistible__bu --------------------------------------------------
MAV_1976|M.avium_104 --------------------------------------------------
MSMEG_1114|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_5243|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1012|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_3561|M.marinum_M --------------------------------------------------
MUL_2794|M.ulcerans_Agy99 --------------------------------------------------
Mb1896c|M.bovis_AF2122/97 --------------------------------------------------
Rv1865c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LRRPWRPRTFARAVSQLFRLTYMVLFSIPVLVPMVLWIALSSATIRRTMV
MAB_3506c|M.abscessus_ATCC_199 --------------------------------------------------
TH_4244|M.thermoresistible__bu --------------------------------------------------
MAV_1976|M.avium_104 --------------------------------------------------
MSMEG_1114|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_5243|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1012|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_3561|M.marinum_M --------------------------------------------------
MUL_2794|M.ulcerans_Agy99 --------------------------------------------------
Mb1896c|M.bovis_AF2122/97 --------------------------------------------------
Rv1865c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DNISTDQIHHSATLARDAAHSVKTYPANYFRAFSGRRRGKGVPVVNVPVQ
MAB_3506c|M.abscessus_ATCC_199 --------------------------------------------------
TH_4244|M.thermoresistible__bu --------------------------------------------------
MAV_1976|M.avium_104 --------------------------------------------------
MSMEG_1114|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_5243|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1012|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_3561|M.marinum_M --------------------------------------------------
MUL_2794|M.ulcerans_Agy99 --------------------------------------------------
Mb1896c|M.bovis_AF2122/97 --------------------------------------------------
Rv1865c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LIVNTMDRYVRPYGYDATARWVPRLWRRDIKAGHFSPMSHPQVMAAAVHD
MAB_3506c|M.abscessus_ATCC_199 --------------------------------------------------
TH_4244|M.thermoresistible__bu --------------------------------------------------
MAV_1976|M.avium_104 --------------------------MPKTFFITGVSSGLGRALAHGALE
MSMEG_1114|M.smegmatis_MC2_155 -------------------------MSEKVWFITGASRGFGREWAIAALD
Mflv_5243|M.gilvum_PYR-GCK -----------------------MAT----VLITGCSSGYGRATALKFLA
Mvan_1012|M.vanbaalenii_PYR-1 -----------------------MTT----VLITGCSSGYGLATAHRFLD
MMAR_3561|M.marinum_M -----------------------MTSTPPVALITGVSSGIGAAIAVRLAS
MUL_2794|M.ulcerans_Agy99 -----------------------MTSTPPVALITGVSSGIGAAIAVRLAS
Mb1896c|M.bovis_AF2122/97 -----------MPGRTSIGVKIRDKVQDKVIAITGGARGIGLATAAALHN
Rv1865c|M.tuberculosis_H37Rv -----------MPGRTSIGVKIRDKVQDKVIAITGGARGIGLATAAALHN
MLBr_00862|M.leprae_Br4923 LADLVDGKQPSRALLRAQVGRSRDTFGDTLVSVTGAGSGIGRETALALAR
MAB_3506c|M.abscessus_ATCC_199 --------------------------MSKKILITGASSGFGRGAAIELAR
TH_4244|M.thermoresistible__bu --------------------------------VTGAARGIGRSIVTQLAD
:** . * * .
MAV_1976|M.avium_104 AGHTVIGTVRKTADLAPFEALDAK---AHG-RILDVTDHAAVIDTVAEAE
MSMEG_1114|M.smegmatis_MC2_155 RGDKVAATARDTATLADLADKYGD---ALLPLQLDVDDRAADFAAVKQAH
Mflv_5243|M.gilvum_PYR-GCK EGWNVVATMRNPRPDV---LPASD---RLRVSALDVTDPESIASAV----
Mvan_1012|M.vanbaalenii_PYR-1 EGWNVVATMRTPRQDV---LPPSD---RLRVIALDVTRPDSIAAAV----
MMAR_3561|M.marinum_M AGFRVVGTSRAPQR-----LAPIP---GVETLALDVTDDTAVRSVVSEVI
MUL_2794|M.ulcerans_Agy99 AGFRVVGTSRAPQR-----LAPIP---GVETLALDVTDDTAVRSVVSEVI
Mb1896c|M.bovis_AF2122/97 LGAKVAIGDIDEAMAKE-SGADLD---LDMYGKLDVTDPDSFSGFLDAVE
Rv1865c|M.tuberculosis_H37Rv LGAKVAIGDIDEAMAKE-SGADLD---LDMYGKLDVTDPDSFSGFLDAVE
MLBr_00862|M.leprae_Br4923 RGAEVVVSDIDEAAAKNTAAQIVASGGVAYAYALDVSDAAAVEAFAEQVS
MAB_3506c|M.abscessus_ATCC_199 RGHQVVATAETWPQVRTLRADAAEAGVDLEVIKLNLLDEIDIAHAAG---
TH_4244|M.thermoresistible__bu SGWDVVAGVRTEQDAENIVALNPA---RITSVLLDVTEADHIAALDR---
* * *::
MAV_1976|M.avium_104 ASIGPIDVLIANAGYGLEGVFEETPLSEVRAQFETNFFGAAATVQSVLPY
MSMEG_1114|M.smegmatis_MC2_155 EHFGRLDIVVNNAGYGQFGFIEELSEAEARAQIETNLFGALWVTQAALPI
Mflv_5243|M.gilvum_PYR-GCK EAAGPLDAVVNNAGIGAVGAFEATPMSTIRELFDTNTFGVMAMTQAVVPQ
Mvan_1012|M.vanbaalenii_PYR-1 AAAGPIDVLVNNAGIGAVGAFEATPMQTVRELFDTNTFGVMAMTQAVVPQ
MMAR_3561|M.marinum_M DRTGRIDVLVNNAGLGIAGAAEESSIDQARSLFDTNFFGLIRLTNEVLPH
MUL_2794|M.ulcerans_Agy99 DRTGHIDVLVNNAGLGIAGAAEESSIDQARSLFDTNFFGLIRLTNEVLPH
Mb1896c|M.bovis_AF2122/97 RQLGPIDVLVNNAGIMPVGRIVDEPDPVTRRILDINVYGVILGSKLAAQR
Rv1865c|M.tuberculosis_H37Rv RQLGPIDVLVNNAGIMPVGRIVDEPDPVTRRILDINVYGVILGSKLAAQR
MLBr_00862|M.leprae_Br4923 AKHGVPDIVVNNAGIGQAGRFLDTPPEEFDRVLAVNLGGVVNCCRAFGQR
MAB_3506c|M.abscessus_ATCC_199 ---YDPDVLVLNAGVMESGSIIDIPLARVRESFDINVFGHIQLAQGIVPK
TH_4244|M.thermoresistible__bu SLDGRLDAVINNAGVVVPGPLETVSPQDLRRQFDVNVIGHLAVTQAVLPR
* :: *** * . : * * .
MAV_1976|M.avium_104 MRKRRGG-HIMAVTSMGGLMTVPGLSVYCASKYALEGLLESIGKEVAGFG
MSMEG_1114|M.smegmatis_MC2_155 LRAQRSG-HIIQVSSIGGISAFPLVGMYHASKWALEGFSQSLAQEVEGFG
Mflv_5243|M.gilvum_PYR-GCK MRERRSG-VVVNVTSSTTLAPMPLAAAYTASKSAIEGFTGSLALELGFFG
Mvan_1012|M.vanbaalenii_PYR-1 MRARRSG-VVVNVTSSVTLAAMPLAAAYTASKAAIEGFTGSLALELGFFG
MMAR_3561|M.marinum_M MRRRGSG-RIINISSVLGFLPAPYAALYAASKHAVEGYTESLDHELREYG
MUL_2794|M.ulcerans_Agy99 MRRRGSG-RIINISSVLGFLPAPYAALYAASKHAVEGYTESLDHELREYG
Mb1896c|M.bovis_AF2122/97 MVPRGRG-HVINVASLAGEIYAVGVATYCASKHAVVAFTDSARLEYRSAG
Rv1865c|M.tuberculosis_H37Rv MVPRGRG-HVINVASLAGEIYAVGVATYCASKHAVVAFTDSARLEYRSAG
MLBr_00862|M.leprae_Br4923 LVERGTGGHIVNVSSMAAYAPLQSLSAYCTSKAATYMFSDCLRAELDAVG
MAB_3506c|M.abscessus_ATCC_199 MVARKSG-KVVWTSSMGGILVIPFVGVYCATKHAIEAIAGSMKAELEPHG
TH_4244|M.thermoresistible__bu LRDSRGR--IVFISSLNGRISVPLMGAYCASKFALEAVADALRMELAPWR
: :: :* . * ::* * . *
MAV_1976|M.avium_104 IHVTAIEPGSFR-TDWAGRSMVRAERAIDDYDAVFEPIRSARL-------
MSMEG_1114|M.smegmatis_MC2_155 INVTLIEPGGFS-TDWAG-SSAKHATPIADYDELRDATNTARS-------
Mflv_5243|M.gilvum_PYR-GCK VRAKLVEPGYGPGTQFTSNGVERMAGLIPAEYQDFAAPVFEEF-------
Mvan_1012|M.vanbaalenii_PYR-1 VRAKLVEPGYGPGTQFASNGSHRMADLIPDEYQDFAAPIMESF-------
MMAR_3561|M.marinum_M VRALLVEPSYTR-TDFESN--MWEADTPQAAYADNRAAVRAVV-------
MUL_2794|M.ulcerans_Agy99 VRALLVEPSYTR-TDFESN--MWEADTPQAAYADNRAAVRAVV-------
Mb1896c|M.bovis_AF2122/97 VKFSMVLPSFVN-TELIAG-----TGGIKGFKNAEPADIADAI-------
Rv1865c|M.tuberculosis_H37Rv VKFSMVLPSFVN-TELIAG-----TGGIKGFKNAEPADIADAI-------
MLBr_00862|M.leprae_Br4923 VGLTTICPGVIDTNIIQSTR----IDTPTAKQERVRDRRGQLD-------
MAB_3506c|M.abscessus_ATCC_199 VKVATVNPGLFG-TGFNDTG---AESHVQWYDPERAVVPMPSF-------
TH_4244|M.thermoresistible__bu IPVIVIQPTQTD-TDMVRGADVMVEQTAAAMRPEHRDLYARHIGGMRKFI
: : * .
MAV_1976|M.avium_104 RANGNQLGDPAKAATAILTAVEAAQPPRHLLLGSDALRLIRAGRDAVDAD
MSMEG_1114|M.smegmatis_MC2_155 QRN-SKPGDPKASASAVLKIVDAEKPPLRVFFGERPLEIAKAD---YERR
Mflv_5243|M.gilvum_PYR-GCK RSPG-MVTTPDEVADVVFQAATDTSGRLRFAAGADAVALAQ---------
Mvan_1012|M.vanbaalenii_PYR-1 GSPA-AVTTPDDVAEVVYLAATDASDRLRYAAGADAVALAQ---------
MMAR_3561|M.marinum_M ADKIRNAELPAVVADATYAAATDKRIKLRYPAGRHARQLALLKRIAPAAV
MUL_2794|M.ulcerans_Agy99 ANKIRNAELPAVVADATYAAAIDKRIKLRYPAGRHARQLALLKRIAPAAV
Mb1896c|M.bovis_AF2122/97 VGLIVHPKPRVRVTKAAGSMIVAQRFMPRQVSEGLNRLLGGEHVFTDDVD
Rv1865c|M.tuberculosis_H37Rv VGLIVHPKPRVRVTKAAGSMIVAQRFMPRQVSEGLNRLLGGEHVFTDDVD
MLBr_00862|M.leprae_Br4923 MMFRLRRYGPDKVADTILSAIKKNKPIRPVAPEAYALYGISRVLPQVLRS
MAB_3506c|M.abscessus_ATCC_199 GDTLADQFDPQEMIDAMVEIIPADEHLYRTMRPLATIDVAKGWQETEWTQ
TH_4244|M.thermoresistible__bu RQFHPSAVPADNVAAAVVNALTVRRPRSRYIVGITHQFPRLQAIILPNLP
.
MAV_1976|M.avium_104 IAAWESLSRSTDFPDGHQIAAS-
MSMEG_1114|M.smegmatis_MC2_155 IKLWEEWQPVAVEAQG-------
Mflv_5243|M.gilvum_PYR-GCK -----------------------
Mvan_1012|M.vanbaalenii_PYR-1 -----------------------
MMAR_3561|M.marinum_M LDNGIRKQTGLNSPRPTPVLAER
MUL_2794|M.ulcerans_Agy99 LDNGIRKQTGLNSPRPTPVLAER
Mb1896c|M.bovis_AF2122/97 MEKRRTYEARARGEE--------
Rv1865c|M.tuberculosis_H37Rv MEKRRTYEARARGEE--------
MLBr_00862|M.leprae_Br4923 TARIRVI----------------
MAB_3506c|M.abscessus_ATCC_199 NA---------------------
TH_4244|M.thermoresistible__bu PAARDPLLRRLGSQP--------