For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTSLAERTALVTGANRGMGREYVAQLLGRKVAKVYAATRNPLAIDVSDPRVIPLQLDVTDAVSVAEAADL ATDVGILINNAGISRASSVLDKDTSALRGELETNLFGPLALASAFADRIAERSGAIVNVSSVLAWLPLGM SYGVSKAAMWSATESMRIELAPRGVQVVGVYVGLVDTDMGRFADAPKSDPADVVRQVLDGIEAGKEDVLA DEMSRQVRASLNVPARERIARLMGN
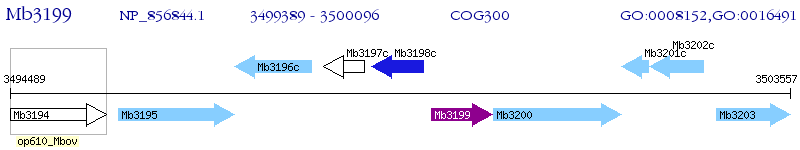
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3199 | - | - | 100% (235) | short chain dehydrogenase |
| M. bovis AF2122 / 97 | Mb2153c | - | 3e-17 | 32.64% (193) | short chain dehydrogenase |
| M. bovis AF2122 / 97 | Mb2237c | ephD | 2e-15 | 35.33% (184) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4104 | - | 2e-17 | 35.64% (188) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv3174 | - | 1e-126 | 99.57% (235) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_00862 | ephD | 9e-17 | 37.50% (184) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4428 | - | 2e-40 | 38.30% (235) | short chain dehydrogenase |
| M. marinum M | MMAR_1614 | - | 1e-18 | 35.91% (181) | short chain dehydrogenase |
| M. avium 104 | MAV_3670 | - | 2e-17 | 32.11% (190) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6581 | - | 1e-50 | 47.47% (217) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_4244 | - | 3e-16 | 32.81% (192) | PUTATIVE putative Dehydrogenase/decarboxylase protein |
| M. ulcerans Agy99 | MUL_3423 | - | 2e-18 | 35.36% (181) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0234 | - | 2e-51 | 48.67% (226) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_6581|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0234|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4428|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_1614|M.marinum_M --------------------------------------------------
MUL_3423|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 MPATQSIHKVPHQFVESPDGVRLAIYQSG--QPEGPAIVLVHGFPDSHVL
MAV_3670|M.avium_104 MTVTSTIT--------ADDGVTLAVHRYTDIDPARPTILAIHGFPDNHHV
TH_4244|M.thermoresistible__bu --------------------------------------------------
Mflv_4104|M.gilvum_PYR-GCK --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_6581|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0234|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4428|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_1614|M.marinum_M --------------------------------------------------
MUL_3423|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 WDGVVPLLAKR-FRIVRYDNRGVGQSSVPKPVSAYTMTRFADDFAAVIDE
MAV_3670|M.avium_104 WDGVADELTGAPYNFVAYDVRGAGESSCPATRSGYGFAQLARDMGAVIDS
TH_4244|M.thermoresistible__bu --------------------------------------------------
Mflv_4104|M.gilvum_PYR-GCK --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_6581|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0234|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4428|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_1614|M.marinum_M --------------------------------------------------
MUL_3423|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LSPDRPVHVLAHDWGSVGVWEYLSRSGAS-----DRVASFTSVSGPSQDQ
MAV_3670|M.avium_104 LGVGR-VHLLAHDWGSIQAWAAVTDQAIMGESVMDKVASFTSISGPHLN-
TH_4244|M.thermoresistible__bu --------------------------------------------------
Mflv_4104|M.gilvum_PYR-GCK --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_6581|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0234|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4428|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_1614|M.marinum_M --------------------------------------------------
MUL_3423|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LVDYILSGLRRPWRPRTFARAVSQLFRLTYMVLFSIPVLVPMVLWIALSS
MAV_3670|M.avium_104 ---YAGAFLRSARTPRAVARVVRQLIASGYIGFFLCPGAAELSFRAGIGV
TH_4244|M.thermoresistible__bu --------------------------------------------------
Mflv_4104|M.gilvum_PYR-GCK --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_6581|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0234|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4428|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_1614|M.marinum_M --------------------------------------------------
MUL_3423|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 ATIRR-TMVDNIST-DQIHHSATLARDAAHSVKTYPANYFRAFSGRRRGK
MAV_3670|M.avium_104 KVIAALERIGHTSTRSQRRDTQRSLRDYLNGLQLYRQNMPAPMLAP--GP
TH_4244|M.thermoresistible__bu --------------------------------------------------
Mflv_4104|M.gilvum_PYR-GCK --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_6581|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0234|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4428|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_1614|M.marinum_M --------------------------------------------------
MUL_3423|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 GVPVVNVPVQLIVNTMDRYVRPYGYDATARWVPRLWRRDIKAGHFSPMSH
MAV_3670|M.avium_104 RLPETTVPVQTLVPRRDVFVTPALQRFTGAIPQRARVIEIEGGHWVVTSR
TH_4244|M.thermoresistible__bu --------------------------------------------------
Mflv_4104|M.gilvum_PYR-GCK --------------------------------------------------
Mb3199|M.bovis_AF2122/97 ----------------------------------MTSLAERTALVTGANR
Rv3174|M.tuberculosis_H37Rv ----------------------------------MTSLAERTVLVTGANR
MSMEG_6581|M.smegmatis_MC2_155 -----------------------------------MKIQGSVAFVTGANR
Mvan_0234|M.vanbaalenii_PYR-1 ----------------------------------MTDTEHTTALVTGANR
MAB_4428|M.abscessus_ATCC_1997 ------------------------------------MIKDTVVLVTGGRR
MMAR_1614|M.marinum_M ---------------------------------------MSTWLITGCST
MUL_3423|M.ulcerans_Agy99 ---------------------------------------MSTWLITGCST
MLBr_00862|M.leprae_Br4923 PQVMAAAVHDLADLVDG--KQPSRALLRAQVGRSRDTFGDTLVSVTGAGS
MAV_3670|M.avium_104 PDVIARLTTEWVDLQSAGAVAPQLTGERRELPGERREVRGRLALVTGAGA
TH_4244|M.thermoresistible__bu --------------------------------------------VTGAAR
Mflv_4104|M.gilvum_PYR-GCK ------------------------------------MAADRIALVTGAAR
:**
Mb3199|M.bovis_AF2122/97 GMGREYVAQLLGRKVAKVYAATRNPLAIDVSDP-------RVIPLQLDVT
Rv3174|M.tuberculosis_H37Rv GMGREYVAQLLGRKVAKVYAATRNPLAIDVSDP-------RVIPLQLDVT
MSMEG_6581|M.smegmatis_MC2_155 GLGRQFAEQLLARG-AKVYAGARNPETVTLDG---------VIPVRIDIT
Mvan_0234|M.vanbaalenii_PYR-1 GLGRRFAEALVARG-AKVYAAARRPETIDIPG---------VVPIQLDIT
MAB_4428|M.abscessus_ATCC_1997 GLGAAIVDEVLNRGARKVYSTARAP--FEDPRA-------QVVTKQLEVT
MMAR_1614|M.marinum_M GLGRALAEAVIDAGHHTVATARSVGGVADLVQRSP----ERVLPLALDIT
MUL_3423|M.ulcerans_Agy99 GLGRALAEAVIDAGHHTVATARSVGGVADLAQRSP----ERVLPLALDIT
MLBr_00862|M.leprae_Br4923 GIGRETALALARRGA-EVVVSDIDEAAAKNTAAQIVASGGVAYAYALDVS
MAV_3670|M.avium_104 GIGRATAVELARQGARKVVIVDRDRAAADQTADAVRAAGAGAAVYQVDVS
TH_4244|M.thermoresistible__bu GIGRSIVTQLADSGWDVVAGVRTEQDAENIVALNP----ARITSVLLDVT
Mflv_4104|M.gilvum_PYR-GCK GQGAAIVTRLLQDGFRVAACDLLGDDVTRTVAAHDT---DHAVAIELDVT
* * . : . ::::
Mb3199|M.bovis_AF2122/97 DA----VSVAEAADLATDVGILINNAGISRASSVLDKDTSALRGELETNL
Rv3174|M.tuberculosis_H37Rv DA----VSVAEAADLATDVGILINNAGISRASSVLDKDTSALRGELETNL
MSMEG_6581|M.smegmatis_MC2_155 DP----ASVQAAAELAADPTILVNNAGTTSYNGLLDGPMDDIRAQMESHF
Mvan_0234|M.vanbaalenii_PYR-1 DP----ESVRRAAEQATDVTVVINNAGVSTRAGLLDGTLEDIRLEMDTHY
MAB_4428|M.abscessus_ATCC_1997 SA----ESVDALARDLTDVEIVVNNAGVLHPDPLLTGDFDRIDATFQTNV
MMAR_1614|M.marinum_M EPDQITAAVQAAQQRFGGIDVLVNNAGYGYRAAVEEGDDAEVRDLFETHF
MUL_3423|M.ulcerans_Agy99 EPDQITAAMQAAQQRFGGIDVLVNNAGYGYRAAVEEGDDAEVRDLFETHF
MLBr_00862|M.leprae_Br4923 DAAAVEAFAEQVSAKHGVPDIVVNNAGIGQAGRFLDTPPEEFDRVLAVNL
MAV_3670|M.avium_104 DEAAMNDLAAQVRNEHGVVDILVNNAGIGMAGRFLETSPANWDAIMGVNV
TH_4244|M.thermoresistible__bu EAD---HIAALDRSLDGRLDAVINNAGVVVPGPLETVSPQDLRRQFDVNV
Mflv_4104|M.gilvum_PYR-GCK SAEQWTRAVGEVVDRFGGLSTLVNNAGVLHRASLADETPDGFEGSWRVNS
. ::**** . :
Mb3199|M.bovis_AF2122/97 FGPLALASAFADRIAERS--GAIVNVSSVLAWLPLG--MSYGVSKAAMWS
Rv3174|M.tuberculosis_H37Rv FGPLALASAFADRIAERS--GAIVNVSSVLAWLPLG--MSYGVSKAAMWS
MSMEG_6581|M.smegmatis_MC2_155 FGTLNVTRAFVPHLVANAP-GAILNIASILSWKHIPGIGAYAAAKAALWA
Mvan_0234|M.vanbaalenii_PYR-1 FGTLAVTRAFVPVIERNGG-GAILNVLSVLSWLHPPTSGAYSAAKAAGWA
MAB_4428|M.abscessus_ATCC_1997 FGPLRIARAFAPILKANGG-GAIVNIHSVLSWLGGA--GAYGASKAAIWS
MMAR_1614|M.marinum_M FGTVALIKAVLPDMRARRS-GAIVNISSIAVALTPVGSGYYAAAKAAMEG
MUL_3423|M.ulcerans_Agy99 FGTVALIKAVLPDMRARRS-GAIVNISSIAVALTPVGSGYYAAAKAAMEG
MLBr_00862|M.leprae_Br4923 GGVVNCCRAFGQRLVERGTGGHIVNVSSMAAYAPLQSLSAYCTSKAATYM
MAV_3670|M.avium_104 RGVIWGSRAFGAQMVERGQGGTIINVASAAAYLPSKSMVAYGTTKAAVLA
TH_4244|M.thermoresistible__bu IGHLAVTQAVLPRLRDSRG--RIVFISSLNGRISVPLMGAYCASKFALEA
Mflv_4104|M.gilvum_PYR-GCK LGPFLGIQAALPHLKAAEG-AAIVNTCSTGAIRPFPNHAAYGSSKWALRG
* . * : *: * * :* *
Mb3199|M.bovis_AF2122/97 ATESMRIELAPRGVQVVGVYVGLVDTDMGRFADAP---------------
Rv3174|M.tuberculosis_H37Rv ATESMRIELAPRGVQVVGVYVGLVDTDMGRFADAP---------------
MSMEG_6581|M.smegmatis_MC2_155 QTDAVREELAPKGVAVTGLYVGFMDTDMAAGVDAE---------------
Mvan_0234|M.vanbaalenii_PYR-1 MTDALRAELAPRGIHVAALHVGFMDTDMVSYIPADQ--------------
MAB_4428|M.abscessus_ATCC_1997 VSNTLRLELEAQHTHVLGVHAAFIDTDMVSALPVP---------------
MMAR_1614|M.marinum_M MSGALHGELAPLGISVTVVEPGAFRTDFAGRSLVQSTRMINDY-------
MUL_3423|M.ulcerans_Agy99 MSGALHGELAPLGISVTVVEPGAFRTDFAGRSLVQSTRMINDY-------
MLBr_00862|M.leprae_Br4923 FSDCLRAELDAVGVGLTTICPGVIDTNIIQSTRIDTPTAKQ---------
MAV_3670|M.avium_104 LSESLRADFADEGITVTAVCPGFVNTNIAKSTIYAGMSAQQQ--------
TH_4244|M.thermoresistible__bu VADALRMELAPWRIPVIVIQPTQTDTDMVRGADVMVEQTAAAMRPEHRDL
Mflv_4104|M.gilvum_PYR-GCK LTQVAAVELAPSGIRVNAVFPGPVETPMLDANTQT---------------
: :: : : * :
Mb3199|M.bovis_AF2122/97 ------------------KSDPADVVRQVLDGIEA---------GKEDVL
Rv3174|M.tuberculosis_H37Rv ------------------KSDPADVVRQVLDGIEA---------GKEDVL
MSMEG_6581|M.smegmatis_MC2_155 ------------------KSDPAHIAAAALDGVEA---------GVAEVL
Mvan_0234|M.vanbaalenii_PYR-1 ------------------KTDPAVVAELALDGLFA---------GQPEIL
MAB_4428|M.abscessus_ATCC_1997 ------------------KTPPSAIAARIIDALDK---------GDNEVL
MMAR_1614|M.marinum_M AATAGQRRKENDTMHGNQPGDPAKAAAAIITAVESPAPPAFLLLGPDALM
MUL_3423|M.ulcerans_Agy99 AATAGQRRKENDTMHGNQPGDPAKAAAAIITAVESPAPPAFLLLGPDALM
MLBr_00862|M.leprae_Br4923 -ERVRDRRGQLDMMFRLRRYGPDKVADTILSAIKKNK-------PIRPVA
MAV_3670|M.avium_104 -ERARDR---ADAAYRRRNFTPEATAKAIVKAIKTGP-------AVVPIA
TH_4244|M.thermoresistible__bu YARHIGGMRKFIRQFHPSAVPADNVAAAVVNALTVRRPRSRYIVGITHQF
Mflv_4104|M.gilvum_PYR-GCK ------------------RLAANALMGRIGKPMEIAD-------AVAFLV
. :
Mb3199|M.bovis_AF2122/97 ADEMSRQVRASLNVPARERIARLMGN--
Rv3174|M.tuberculosis_H37Rv ADEMSRQVRASLNVPARERIARLMGN--
MSMEG_6581|M.smegmatis_MC2_155 GDEQTRAVKASLSA--------------
Mvan_0234|M.vanbaalenii_PYR-1 ADELTRNVKAQLSAAPA-----------
MAB_4428|M.abscessus_ATCC_1997 TDDLTVQIKSQLAGPVEKLAYRPGH---
MMAR_1614|M.marinum_M AYRHIAQRRDVEITDWEQLTAGTNLTP-
MUL_3423|M.ulcerans_Agy99 AYRHIAQRRDVEITDWEQLTAGTNLTP-
MLBr_00862|M.leprae_Br4923 PEAYALYGISRVLPQVLRSTARIRVI--
MAV_3670|M.avium_104 AESRIGYALRRISPGAIRLLARWDIRQT
TH_4244|M.thermoresistible__bu PRLQAIILPNLPPAARDPLLRRLGSQP-
Mflv_4104|M.gilvum_PYR-GCK SEHATFITGSELIVDGGQSLQIG-----