For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DTSALLKSVPTGLWIGGEERQGTSTFSVLDPSDDEVLATVADATADDARDALDAACAVQGEWAATPARKR GEILRSVFETITARADDIAALMTLEMGKVFAESKGEVTYGAEFFRWFAEEAVRIEGRYTPAPAGTGRILV TKQAVGPCYAITPWNFPLAMGTRKMGPAFAAGCTMIVKPAQETPLTMLLLAKLMAEAGLPKGVLSVLPTS NPGAVTEALINDGRLRKLTFTGSTGVGKALVKQSADKLLRTSMELGGNAPFIVFDDADVDAAVDGAILAK MRNGGEACTAANRIHVANAVREEFTEKFVKRMSEFTLGKGLDEKSTLGPLINAKQVATVTELVSDAVSRG ATVAVGGVAPGGPGNFYPATVLTDVPADARILKEEVFGPVAPIAGFDTEEEGIAAANDTEYGLAAYVYTQ SLDRALRVAENIESGMVGVNRGVISDAAAPFGGIKESGFGREGGTEGIEEYLDTKYIALTR*
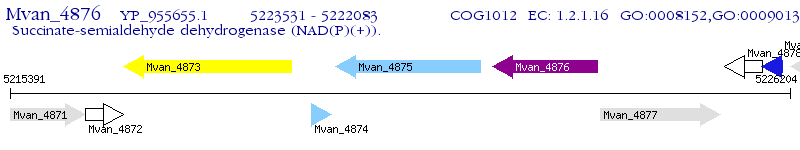
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4876 | - | - | 100% (482) | succinate-semialdehyde dehydrogenase (NAD(P)(+)) |
| M. vanbaalenii PYR-1 | Mvan_0014 | - | 8e-87 | 37.82% (476) | betaine-aldehyde dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0352 | - | 6e-85 | 38.89% (486) | aldehyde dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2883c | aldC | 1e-72 | 35.68% (454) | aldehyde dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1858 | - | 0.0 | 94.02% (485) | succinate-semialdehyde dehydrogenase (NAD(P)(+)) |
| M. tuberculosis H37Rv | Rv2858c | aldC | 1e-72 | 35.68% (454) | aldehyde dehydrogenase |
| M. leprae Br4923 | MLBr_02573 | gabD1 | 2e-74 | 35.38% (455) | succinic semialdehyde dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4208 | - | 0.0 | 69.85% (481) | putative succinate-semialdehyde dehydrogenase |
| M. marinum M | MMAR_3556 | - | 2e-83 | 37.18% (468) | aldehyde dehydrogenase NAD dependent |
| M. avium 104 | MAV_2101 | - | 1e-80 | 37.29% (472) | aldehyde dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5538 | - | 0.0 | 90.99% (477) | [NADP+] succinate-semialdehyde dehydrogenase [Mycobacterium |
| M. thermoresistible (build 8) | TH_3381 | - | 1e-176 | 87.78% (360) | PUTATIVE Succinate-semialdehyde dehydrogenase (NAD(P)(+)) |
| M. ulcerans Agy99 | MUL_2789 | - | 1e-82 | 36.75% (468) | gamma-aminobutyraldehyde dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_4876|M.vanbaalenii_PYR-1 MDTS---ALLKSVPTGLWIGGEERQGTS--TFSVLDPSDDEVLATVADAT
Mflv_1858|M.gilvum_PYR-GCK MDNSAVEALLASVPTGLWIGGEEREGTS--TFDVLDPSDDTVLTSVADAT
MSMEG_5538|M.smegmatis_MC2_155 MRIE---DLISSVPTGLWIGGEEREAAS--TFNVLDPSDDQVITAVADAT
TH_3381|M.thermoresistible__bu --------------------------------------------------
MAB_4208|M.abscessus_ATCC_1997 MTDHS--TLIDAIPTDLWIGGKQVPSSTGTRFPVHNPATGAVLTTVADAA
Mb2883c|M.bovis_AF2122/97 --------MSTTQ--------------------LINPATEEVLASVDHTD
Rv2858c|M.tuberculosis_H37Rv --------MSTTQ--------------------LINPATEEVLASVDHTD
MAV_2101|M.avium_104 --------MSTTIP-------------------VFNPSTEEQIAEVPDSD
MMAR_3556|M.marinum_M --------MTTTVSVAGSWINGAAHSTGGRQHAVINPATQETIAELALAT
MUL_2789|M.ulcerans_Agy99 --------MTTTVSVAGSWINGAAHSTGGRQHAVINPATQETIAELALAT
MLBr_02573|M.leprae_Br4923 -----------------------------MSIATINPATGETVKTFTPTT
Mvan_4876|M.vanbaalenii_PYR-1 ADDARDALDAACAVQGE--WAATPARKRGEILRSVFETITARADDIAALM
Mflv_1858|M.gilvum_PYR-GCK ADDARAALDAACAVQAE--WAGTAPRKRGEILRSVFETITARADDIAALM
MSMEG_5538|M.smegmatis_MC2_155 AEDAIAALDAACAVQDE--WAATPARTRGEILRSVFEKITERADDIAALM
TH_3381|M.thermoresistible__bu -----------------------VVRRRG---------------------
MAB_4208|M.abscessus_ATCC_1997 ASDGAIALDEATAVQRS--WAATAPRERAEILRRAWELVIARRDDFALAM
Mb2883c|M.bovis_AF2122/97 ANAVDDAVQRARAAQRR--WARLAPAQRAAGLRAFAAAVQAHLDELAALE
Rv2858c|M.tuberculosis_H37Rv ANAVDDAVQRARAAQRR--WARLAPAQRAAGLRAFAAAVQAHLDELAALE
MAV_2101|M.avium_104 QAAVDAAVARARETFESGVWRKAPASHRANVLFRAARIIEERKDELAALE
MMAR_3556|M.marinum_M PADVDRAVVAARRALPQ--WAGVTPAERSAVLAKLADLLEQRAPELIAEE
MUL_2789|M.ulcerans_Agy99 PADVDHAVVAARRALPQ--WAGVTTAERSAVLAELADLLEQRAPELIAEE
MLBr_02573|M.leprae_Br4923 QDEVEDAIARAYARFDN--YRHTTFTQRAKWANATADLLEAEANQSAALM
*.
Mvan_4876|M.vanbaalenii_PYR-1 TLEMGKVF-AESKGEVTYGAEFFRWFAEEAVRIEGRYTPAPAG-------
Mflv_1858|M.gilvum_PYR-GCK TLEMGKVL-AESKGEVTYGAEFFRWFAEEAVRIDGRYTAAPAG-------
MSMEG_5538|M.smegmatis_MC2_155 TLEMGKVL-PESKGEVAYGAEFFRWFSEEAVRIDGRYTHAPAG-------
TH_3381|M.thermoresistible__bu ----------------------------------GPPPRAPAG-------
MAB_4208|M.abscessus_ATCC_1997 TLEMGKPL-AESQGEVAYGGEFLRWFSEEAVRINGRYTSSPTG-------
Mb2883c|M.bovis_AF2122/97 VANSGHPI-VSAEWEAGHVRDVLAFYAASPERLSGRQIPVAG----GVDV
Rv2858c|M.tuberculosis_H37Rv VANSGHPI-VSAEWEAGHVRDVLAFYAASPERLSGRQIPVAG----GVDV
MAV_2101|M.avium_104 SQDNGMNL-TAAGHIIPVSVDMLKYYAGWVGKIHGESANLVSDGLLGTWE
MMAR_3556|M.marinum_M VCQTGKPLRLAEEFDVPGSVDNIRFFAGAARRLPGQATAEYS-----ADH
MUL_2789|M.ulcerans_Agy99 VCQTGKPLRLAEEFDVPGSVDNIRFFAGAARRLPGQATAEYS-----ADH
MLBr_02573|M.leprae_Br4923 TLEMGKTL-QSAKDEAMKCAKGFRYYAEKAETLLADEPAEAAAVG-----
.
Mvan_4876|M.vanbaalenii_PYR-1 TGRILVTKQAVGPCYAITPWNFPLAMGTRKMGPAFAAGCTMIVKPAQETP
Mflv_1858|M.gilvum_PYR-GCK TGRILVTKQAVGPCYAITPWNFPLAMGTRKMGPAFAAGCTMIVKPAQETP
MSMEG_5538|M.smegmatis_MC2_155 TGRILVTKQAVGPCYAITPWNFPLAMGTRKMGPAFAAGCTMIVKPAQETP
TH_3381|M.thermoresistible__bu NGRILVTKQAVGPCYAITPWNFPLAMGTRKIGPALAAGCTMIVKPAQETP
MAB_4208|M.abscessus_ATCC_1997 AGRILVAKQPIGPTLAITPWNFPLAMGTRKIGPALAAGCTMIVKPAAETP
Mb2883c|M.bovis_AF2122/97 TFN-----EPMGVVGVITPWNFPMVIASWAIAPALAAGNAVLVKPAELTP
Rv2858c|M.tuberculosis_H37Rv TFN-----EPMGVVGVITPWNFPMVIASWAIAPALAAGNAVLVKPAELTP
MAV_2101|M.avium_104 TYHTFTQLEPVGVVGLIIPWNGPFFVAMLKVAPALAAGCSAVLKPAEETP
MMAR_3556|M.marinum_M TSS--IRREAIGVVATITPWNYPLQMAVWKVMPALAAGCTVVIKPSELTP
MUL_2789|M.ulcerans_Agy99 TSS--IRREAIGVVATITPWNYPLQMAVWKVMPALAAGCTVVIKPSELTP
MLBr_02573|M.leprae_Br4923 ASKALARYQPLGVVLAVMPWNFPLWQTVRFAAPALMAGNVGLLKHASNVP
:.:* : *** *: **: ** ::* : .*
Mvan_4876|M.vanbaalenii_PYR-1 LTMLLLAKLMAEAGLPKGVLSVLPTSNPGAVTEALINDGRLRKLTFTGST
Mflv_1858|M.gilvum_PYR-GCK LTMLLLAKLMADAGLPKGVLSVLPTTSPGPVTETLIDDGRLRKLTFTGST
MSMEG_5538|M.smegmatis_MC2_155 LTMLLLAKLMDEAGLPKGVLSVLPTSNPRGVTGALIDDGRLRKLTFTGST
TH_3381|M.thermoresistible__bu LTMLLLAKLIDEAGLPKGVLSVLPTSNPRPLTEALIGDGRLRKLTFTGST
MAB_4208|M.abscessus_ATCC_1997 LTMLLLGQVFADAGLPAGVLSILPTTNASALTGPLIDDPRLRKLTFTGST
Mb2883c|M.bovis_AF2122/97 LTTMRLGELAVEAGLDEDLLQVLPGKG-TVVGERFVTHPDIRKIVFTGST
Rv2858c|M.tuberculosis_H37Rv LTTMRLGELAVEAGLDEDLLQVLPGKG-TVVGERFVTHPDIRKIVFTGST
MAV_2101|M.avium_104 LTALKLEEIFREAGLPDGVLNVITGYG-ETTGAALTAHPDVDKISFTGST
MMAR_3556|M.marinum_M LTTLTLARLAAEAGLPDGVLNVVTGLG-ADVGAALAAHPGVDLVTFTGST
MUL_2789|M.ulcerans_Agy99 LTTLTLARLAAEAGLPDGVLNVVTGLG-ADVGAALAAHPGVDLVTFTGST
MLBr_02573|M.leprae_Br4923 QCALYLADVIARGGFPEDCFQTLLISA--NGVEAILRDPRVAAATLTGSE
: * : .*: . :. : : . : :***
Mvan_4876|M.vanbaalenii_PYR-1 GVGKALVKQSADKLLRTSMELGGNAPFIVFDDADVDAAVDGAILAKMRNG
Mflv_1858|M.gilvum_PYR-GCK GVGKALVKQSADKLLRTSMELGGNAPFVVFDDADVDAAVDGAILAKMRNG
MSMEG_5538|M.smegmatis_MC2_155 GVGKALVKQSADKLLRTSMELGGNAPFIVFDDADVDAAVDGAILAKMRNG
TH_3381|M.thermoresistible__bu GVGKALAQQASAKLLRTSMELGGNAPFIVFDDADIDAAVDGAMLAKMRNG
MAB_4208|M.abscessus_ATCC_1997 GVGKILLSQCATRVLRSSMELGGNAPFVVFDDADVDAAVDGAMAAKMRNI
Mb2883c|M.bovis_AF2122/97 EVGKRVMAGAAAQVKRVTLELGGKSANIVFHDCDLERAATTAPAGVFDNA
Rv2858c|M.tuberculosis_H37Rv EVGKRVMAGAAAQVKRVTLELGGKSANIVFHDCDLERAATTAPAGVFDNA
MAV_2101|M.avium_104 EVGRLIVKAAAGNLKRLTLELGGKSPLIMFDDANLDKAIMGAGMGLLAGS
MMAR_3556|M.marinum_M AVGRKVMAAAATHGRRTQLELGGKAPFVVFDDADLDAAIQGAVAGALINS
MUL_2789|M.ulcerans_Agy99 AVGRKVMAAAATHGRRTQLELGGKAPFVVFDDADLDAAIQGAVAGAVINS
MLBr_02573|M.leprae_Br4923 PAGQAVGAIAGNEIKPTVLELGGSDPFIVMPSADLDAAVATAVTGRVQNN
.*: : .. . :****. . ::: ..::: * * . . .
Mvan_4876|M.vanbaalenii_PYR-1 GEACTAANRIHVANAVREEFTEKFVKRMSEFTLGKGLDEKSTLGPLINAK
Mflv_1858|M.gilvum_PYR-GCK GEACTAANRIHVANAVREEFTDKFVKRMSEFTLGKGLDEKSTLGPLINAK
MSMEG_5538|M.smegmatis_MC2_155 GEACTAANRFHVANSVREEFTEKLVKRMSEFTLGKGLDPSSTLGPLINSK
TH_3381|M.thermoresistible__bu GEACTAANRFHVANAVREEFTEKLVKRMSEVTLGNGLDGSSTLGPLINAQ
MAB_4208|M.abscessus_ATCC_1997 GEACTAANRFHVDNAVRAEFTEKLTARMGALTIGPGDENGVQVGPLITAK
Mb2883c|M.bovis_AF2122/97 GQDCCARSRILVQRSVYDRFMELLEPAVHSIVVGDPGSRATEMGPLVSRA
Rv2858c|M.tuberculosis_H37Rv GQDCCARSRILVQRSVYDRFMELLEPAVHSIVVGDPGSRATEMGPLVSRA
MAV_2101|M.avium_104 GQNCSCTSRIYVQRGIYDQVVEGLAEFAMMLPMGGSDDPNSVLGPLISEK
MMAR_3556|M.marinum_M GQDCTAATRAIVARELYHDFVAGVADVMSTVVIGDPMDPDTDIGPLISLA
MUL_2789|M.ulcerans_Agy99 GQDCTAATRAIVARELCHDFVAGVADVMSTVVIGDPMDPDTDIGPLISLA
MLBr_02573|M.leprae_Br4923 GQSCIAAKRFIAHADIYDEFVEKFVARMETLRVGDPTDPDTDVGPLATES
*: * . .* . : . . . :* . :*** .
Mvan_4876|M.vanbaalenii_PYR-1 QVATVTELVSDAVSRGATVAVGGVAPGGPGNFYPATVLTDVPADARILKE
Mflv_1858|M.gilvum_PYR-GCK QVATVTELVSDAVSRGATVAVGGVAPGGPGNFYPATVLTDVPADARILKE
MSMEG_5538|M.smegmatis_MC2_155 QVATVTELVSDAVSRGATVAVGGVAPGGPGNFYPATVLADVPADARILKE
TH_3381|M.thermoresistible__bu QRDTVVELVTDAVERGATVAVGGTVPDGPGWFYPATVLTDVSADARILKE
MAB_4208|M.abscessus_ATCC_1997 QRASVDELVQDAVSKGAVVTTGGVVADGEGYFYPPTVLTDVPADARILRE
Mb2883c|M.bovis_AF2122/97 HRDKVAGYVPD----DAPVAFRGTAPAGRGFWFPPTVLTPKRGD-RTVTD
Rv2858c|M.tuberculosis_H37Rv HRDKVAGYVPD----DAPVAFRGTAPAGRGFWFPPTVLTPKRGD-RTVTD
MAV_2101|M.avium_104 QRQRVEGIVNDGVAAGAKVITGGKPMDRKGYFYEATIITNTTPDMRLIRE
MMAR_3556|M.marinum_M HRDRVADIVAKAPAQGARIVVGGRIPDLAGAFYLPTLLADVGEQSQAYRE
MUL_2789|M.ulcerans_Agy99 HRDRVADILAKAPAQGARIVVGGRIPDLAGAFYLPTLLADVGEQSQAYRE
MLBr_02573|M.leprae_Br4923 GRAQVEQQVDAAAAAGAVIRCGGKRPDRPGWFYPPTVITDITKDMALYTD
* : .* : * * :: .*::: : :
Mvan_4876|M.vanbaalenii_PYR-1 EVFGPVAPIAGFDTEEEGIAAANDTEYGLAAYVYTQSLDRALRVAENIES
Mflv_1858|M.gilvum_PYR-GCK EVFGPVAPIAGFDTEEEGIAAANDTEYGLAAYVYTQSLDRALRVAEGIES
MSMEG_5538|M.smegmatis_MC2_155 EVFGPVAPITGFDTEEEGIAAANDTEYGLAAYIYTQSLDRALRVAETIQS
TH_3381|M.thermoresistible__bu EVFGPVAPIVGFDTEEEGIAAANDTEYGLAAYIYTRSLDRALRVAEAIES
MAB_4208|M.abscessus_ATCC_1997 EVFGPVAAITGFDGEEAGIAAANDTEFGLAAYVYTANLDRALRVSQAVES
Mb2883c|M.bovis_AF2122/97 EIFGPVVVVLTFDDEADAISLANDTAYGLSGSIWTDDLSRALRVARAVES
Rv2858c|M.tuberculosis_H37Rv EIFGPVVVVLTFDDEADAISLANDTAYGLSGSIWTDDLSRALRVARAVES
MAV_2101|M.avium_104 EIFGPVGCVIPFDDEDEVIAAANDTHYGLAGAIWTENLSRAHRVVNELRA
MMAR_3556|M.marinum_M EIFGPVLTVRAHDGDDDALRQANDTDYGLAASAWTRDVYRAQRASREISA
MUL_2789|M.ulcerans_Agy99 EIFGPVLTVRAHDGDDDALRQANDTDYGLAASAWTRDVYRAQRASREISA
MLBr_02573|M.leprae_Br4923 EVFGPVASVYCATNIDDAIEIANATPFGLGSNAWTQNESEQRHFIDNIVA
*:**** : : ** * :**.. :* . . : : :
Mvan_4876|M.vanbaalenii_PYR-1 GMVGVNRGVISDAAAPFGGIKESGFGREGGTEGIEEYLDTKYIALTR---
Mflv_1858|M.gilvum_PYR-GCK GMVGVNRGVISDPAAPFGGIKESGFGREGGTEGIEEYLDTKYIALTK---
MSMEG_5538|M.smegmatis_MC2_155 GMVGINRGVISDPAAPFGGIKESGFGREGGIEGIEEYLDTKYIALTK---
TH_3381|M.thermoresistible__bu GMVGVNRGVISDAAAPFGGIKESGFGREGGTEGIEEYLETKYIALT----
MAB_4208|M.abscessus_ATCC_1997 GMVGVNRGVISDVAAPFGGIKESGIGREAGSEGIEEYLETKYIALP----
Mb2883c|M.bovis_AF2122/97 GNLSVNSHSSVRFNTPFGGFKQSGVGRELGPDAPLQFTETKNVFIAVGEE
Rv2858c|M.tuberculosis_H37Rv GNLSVNSHSSVRFNTPFGGFKQSGVGRELGPDAPLQFTETKNVFIAVGEE
MAV_2101|M.avium_104 GQIWVNSALAADPSMPISGHKQSGWGGERGRKGIEAYFNTKAVYIGL---
MMAR_3556|M.marinum_M GCVWINDHIPIVSEMPHGGVAASGFGKDMSEYSFEEYLTIKHVMSDITGV
MUL_2789|M.ulcerans_Agy99 GCVWINDHIPIVSEMPHGGVAASGFGNDMSEYSFDEYLTIKHVMSDITGV
MLBr_02573|M.leprae_Br4923 GQVFINGMTVSYPELPFGGIKRSGYGRELSTHGIREFCNIKTVWIA----
* : :* * .* ** * : . . : * :
Mvan_4876|M.vanbaalenii_PYR-1 --------------
Mflv_1858|M.gilvum_PYR-GCK --------------
MSMEG_5538|M.smegmatis_MC2_155 --------------
TH_3381|M.thermoresistible__bu --------------
MAB_4208|M.abscessus_ATCC_1997 --------------
Mb2883c|M.bovis_AF2122/97 M-------------
Rv2858c|M.tuberculosis_H37Rv M-------------
MAV_2101|M.avium_104 --------------
MMAR_3556|M.marinum_M AEKPWHRTIFSTRI
MUL_2789|M.ulcerans_Agy99 AEKPWHRTIFSTRI
MLBr_02573|M.leprae_Br4923 --------------