For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TTTVSVAGSWINGAAHSTGGRQHAVINPATQETIAELALATPADVDRAVVAARRALPQWAGVTPAERSAV LAKLADLLEQRAPELIAEEVCQTGKPLRLAEEFDVPGSVDNIRFFAGAARRLPGQATAEYSADHTSSIRR EAIGVVATITPWNYPLQMAVWKVMPALAAGCTVVIKPSELTPLTTLTLARLAAEAGLPDGVLNVVTGLGA DVGAALAAHPGVDLVTFTGSTAVGRKVMAAAATHGRRTQLELGGKAPFVVFDDADLDAAIQGAVAGALIN SGQDCTAATRAIVARELYHDFVAGVADVMSTVVIGDPMDPDTDIGPLISLAHRDRVADIVAKAPAQGARI VVGGRIPDLAGAFYLPTLLADVGEQSQAYREEIFGPVLTVRAHDGDDDALRQANDTDYGLAASAWTRDVY RAQRASREISAGCVWINDHIPIVSEMPHGGVAASGFGKDMSEYSFEEYLTIKHVMSDITGVAEKPWHRTI FSTRI*
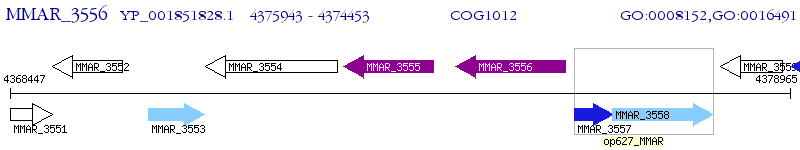
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3556 | - | - | 100% (496) | aldehyde dehydrogenase NAD dependent |
| M. marinum M | MMAR_2610 | - | e-120 | 48.39% (467) | aldehyde dehydrogenase |
| M. marinum M | MMAR_2875 | - | 1e-98 | 39.68% (499) | aldehyde dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0791 | aldA | 3e-87 | 42.45% (457) | aldehyde dehydrogenase NAD dependant AldA |
| M. gilvum PYR-GCK | Mflv_4860 | - | 0.0 | 78.00% (491) | gamma-aminobutyraldehyde dehydrogenase |
| M. tuberculosis H37Rv | Rv0768 | aldA | 3e-87 | 42.45% (457) | aldehyde dehydrogenase NAD dependent AldA |
| M. leprae Br4923 | MLBr_02573 | gabD1 | 6e-72 | 36.20% (453) | succinic semialdehyde dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3328 | - | 0.0 | 79.02% (491) | gamma-aminobutyraldehyde dehydrogenase |
| M. avium 104 | MAV_2608 | - | 3e-96 | 40.67% (477) | aldehyde dehydrogenase family protein |
| M. smegmatis MC2 155 | MSMEG_1665 | - | 0.0 | 79.43% (491) | gamma-aminobutyraldehyde dehydrogenase |
| M. thermoresistible (build 8) | TH_3080 | - | 5e-89 | 41.63% (478) | PUTATIVE aldehyde dehydrogenase |
| M. ulcerans Agy99 | MUL_2789 | - | 0.0 | 98.39% (496) | gamma-aminobutyraldehyde dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_1572 | - | 0.0 | 78.12% (489) | gamma-aminobutyraldehyde dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0791|M.bovis_AF2122/97 ---MALWGDGISALLIDGKLSD-GRAGTFPTVNPATEEVLGVAADADAED
Rv0768|M.tuberculosis_H37Rv ---MALWGDGISALLIDGKLSD-GRAGTFPTVNPATEEVLGVAADADAED
TH_3080|M.thermoresistible__bu --------VHPEELFIGGEWATPSSAQRIEVISPHTEEPVAAVAAACPDD
MAV_2608|M.avium_104 MTDTATETLVQFDLLIGGESSSAASGLTYDSVDPFTGRPWARVPNAAAAD
MMAR_3556|M.marinum_M ----MTTTVSVAGSWING-AAHSTGGRQHAVINPATQETIAELALATPAD
MUL_2789|M.ulcerans_Agy99 ----MTTTVSVAGSWING-AAHSTGGRQHAVINPATQETIAELALATPAD
Mflv_4860|M.gilvum_PYR-GCK --------MTVASSWIDG-APVKTGGAQHTVINPATGESVAEFALAQPGD
Mvan_1572|M.vanbaalenii_PYR-1 ---------MLAGSWIDG-APVNTRGPVHQVINPATGAPVAEYALAQPAD
MSMEG_1665|M.smegmatis_MC2_155 ------MAVKVASSWING-ASVETSGETHQIIDPATGKSVSEYNLATAAD
MAB_3328|M.abscessus_ATCC_1997 --------MSVPSSWING-RPVATHGDTHRVINPATGEAVADLKLATTAD
MLBr_02573|M.leprae_Br4923 --------------------------MSIATINPATGETVKTFTPTTQDE
:.* * : :
Mb0791|M.bovis_AF2122/97 MGRAIEAARRAFDSTDWSR-NTELRVRCVRQLRDAMQQHVEELRELTISE
Rv0768|M.tuberculosis_H37Rv MGRAIEAARRAFDSTDWSR-NTELRVRCVRQLRDAMQQHVEELRELTISE
TH_3080|M.thermoresistible__bu VDRAVAAARAAFDTGPWPRMDPAERVAVIRGLAERYGERRSELAELITAQ
MAV_2608|M.avium_104 VDRAVGAARAALN-GPWGALTATARGKLLHRLGEIIAREADQLAELEVRD
MMAR_3556|M.marinum_M VDRAVVAARRALP--QWAGVTPAERSAVLAKLADLLEQRAPELIAEEVCQ
MUL_2789|M.ulcerans_Agy99 VDHAVVAARRALP--QWAGVTTAERSAVLAELADLLEQRAPELIAEEVCQ
Mflv_4860|M.gilvum_PYR-GCK VDRAVVSARAALP--GWKGATPAERSAVLAKLAKLADDATADLVAEEVSQ
Mvan_1572|M.vanbaalenii_PYR-1 VDAAVASARAALG--GWATATPAERSAVLAKLAKLADEATADLVAEEVSQ
MSMEG_1665|M.smegmatis_MC2_155 VDTAVAAARAAFP--AWAGATPAERSAVLAKLAKLADEHAEQFVAEEVSQ
MAB_3328|M.abscessus_ATCC_1997 VDAAVAAARAAGP--AWASSTPVDRSSVLAKLAAVLSANADTLVADEVAQ
MLBr_02573|M.leprae_Br4923 VEDAIARAYARFD--NYRHTTFTQRAKWANATADLLEAEANQSAALMTLE
: *: * : * :
Mb0791|M.bovis_AF2122/97 VGAPRMLTASAQLEGPVGDLSFAADTAESYPWKQDLGEASPLGIATRRTL
Rv0768|M.tuberculosis_H37Rv VGAPRMLTASAQLEGPVGDLSFAADTAESYPWKQDLGEASPLGIATRRTL
TH_3080|M.thermoresistible__bu IGAPISFAQRAQVGLPWTMMSAFCTLAENHPWQQTRAGMYGADVHVR---
MAV_2608|M.avium_104 GGK-LVREMVGQMRSLPDYYFYYGGLADKLQGDVIPVDKPNYFVYTR---
MMAR_3556|M.marinum_M TGKPLRLAEEFDVPGSVDNIRFFAGAARRLPGQATAEYSADHTSSIR---
MUL_2789|M.ulcerans_Agy99 TGKPLRLAEEFDVPGSVDNIRFFAGAARRLPGQATAEYSADHTSSIR---
Mflv_4860|M.gilvum_PYR-GCK TGKPVRLATEFDVPGSVDNIDFFAGAARHLEGKASAEYSGDHTSSIR---
Mvan_1572|M.vanbaalenii_PYR-1 TGKPVRLATEFDVPGSVDNIDFFAGAARHLEGKASGEYSADHTSSIR---
MSMEG_1665|M.smegmatis_MC2_155 TGKPVRLATEFDVPGSVDNIDFFAGAARHLEGKATAEYSGDHTSSIR---
MAB_3328|M.abscessus_ATCC_1997 TGKPVRLASEFDVPGSIDNVEFFAGAARHLEGKATAEYSADHTSSIR---
MLBr_02573|M.leprae_Br4923 MGKTLQSAKDEAMKCAKGFRYYAEKAETLLADEPAEAAAVGASKALAR--
* : .
Mb0791|M.bovis_AF2122/97 AREAVGVVGAITPWNFPHQINLAKLGPALAAGNTVVLKPAPDTPWCAAAL
Rv0768|M.tuberculosis_H37Rv AREAVGVVGAITPWNFPHQINLAKLGPALAAGNTVVLKPAPDTPWCAAAL
TH_3080|M.thermoresistible__bu -REPVGVVAAIVPWNMPQFLIVTKLIPALLAGCTVVLKPAPESPLDALLL
MAV_2608|M.avium_104 -HEPVGVVGAITPWNSPLLLLTWKLAAGLAAGCTFVVKPSDHTPTSTLAF
MMAR_3556|M.marinum_M -REAIGVVATITPWNYPLQMAVWKVMPALAAGCTVVIKPSELTPLTTLTL
MUL_2789|M.ulcerans_Agy99 -REAIGVVATITPWNYPLQMAVWKVMPALAAGCTVVIKPSELTPLTTLTL
Mflv_4860|M.gilvum_PYR-GCK -REAVGVVATITPWNYPLQMAVWKVIPALAAGCSVVIKPAEITPLTTLTL
Mvan_1572|M.vanbaalenii_PYR-1 -REAVGVVATITPWNYPLQMAVWKVLPALAAGCSVVIKPAEITPLTTLTL
MSMEG_1665|M.smegmatis_MC2_155 -REAVGVVATITPWNYPLQMAVWKVIPALAAGCAVVIKPAEITPLTTLTL
MAB_3328|M.abscessus_ATCC_1997 -REAAGVVATITPWNYPLQMAVWKVLPALAAGCTVVIKPSELTPLTTLTL
MLBr_02573|M.leprae_Br4923 -YQPLGVVLAVMPWNFPLWQTVRFAAPALMAGNVGLLKHASNVPQCALYL
:. *** :: *** * ..* ** ::* : * : :
Mb0791|M.bovis_AF2122/97 GEIIVEHTDFPPGVVNIVTSSSHALGALLAKDPRVDMISFTGSTATGRAV
Rv0768|M.tuberculosis_H37Rv GEIIVEHTDFPPGVVNIVTSSSHALGALLAKDPRVDMISFTGSTATGRAV
TH_3080|M.thermoresistible__bu AELIADS-GIPSGVVNVLPG-PAGVGERLVRHPGVDKVSFTGSTAAGRAV
MAV_2608|M.avium_104 AKLFAEA-GFPPGVINVVTGWGPETGAALASHPGVDKVAFTGSTATGIHV
MMAR_3556|M.marinum_M ARLAAEA-GLPDGVLNVVTGLGADVGAALAAHPGVDLVTFTGSTAVGRKV
MUL_2789|M.ulcerans_Agy99 ARLAAEA-GLPDGVLNVVTGLGADVGAALAAHPGVDLVTFTGSTAVGRKV
Mflv_4860|M.gilvum_PYR-GCK ARLASEA-GLPDGVLNVVTGSGADVGAALAGHADVDMVTFTGSTPVGRRV
Mvan_1572|M.vanbaalenii_PYR-1 ARLAAEA-GLPAGVFNVVTGSGADVGTALAGHADVDMVTFTGSTPVGRKV
MSMEG_1665|M.smegmatis_MC2_155 ARLASEA-GLPDGVFNVVTGSGAEVGTALAGHRDVDVVTFTGSTAVGRKV
MAB_3328|M.abscessus_ATCC_1997 ARLAAEA-GLPEGVLNVVTGRGDDVGYALASHPDVDLVTFTGSTAVGRKV
MLBr_02573|M.leprae_Br4923 ADVIARG-GFPEDCFQTLLISANGVEAILR-DPRVAAATLTGSEPAGQAV
. : .:* . .: : * . * ::*** ..* *
Mb0791|M.bovis_AF2122/97 MADAAATIKKVFLELGGKSAFVVLDDADLAAASAVSAFSACMHAGQGCAI
Rv0768|M.tuberculosis_H37Rv MADAAATIKKVFLELGGKSAFVVLDDADLAAASAVSAFSACMHAGQGCAI
TH_3080|M.thermoresistible__bu AAAAAAGLKRVSLELGGKSAAVVLSDADPAAVATAVRSASLSNSGQICNA
MAV_2608|M.avium_104 GKAAIENMTRFSLELGGKSAQVVFADADLDAAANGVIAGVFAATGQTCLA
MMAR_3556|M.marinum_M MAAAATHGRRTQLELGGKAPFVVFDDADLDAAIQGAVAGALINSGQDCTA
MUL_2789|M.ulcerans_Agy99 MAAAATHGRRTQLELGGKAPFVVFDDADLDAAIQGAVAGAVINSGQDCTA
Mflv_4860|M.gilvum_PYR-GCK MLAAAAHGHRTQLELGGKAPFVVFDDADLDAAIQGAVAGALINTGQDCTA
Mvan_1572|M.vanbaalenii_PYR-1 MLAAAAHGHRTQLELGGKAPFVVFDDADLDAAIQGAVAGALINTGQDCTA
MSMEG_1665|M.smegmatis_MC2_155 MASAAVHGHRTQLELGGKAPFVVFSDADLDAAVQGAVAGALINTGQDCTA
MAB_3328|M.abscessus_ATCC_1997 MAAAAVHGHRTQLELGGKAPFVVFDDADMDAAIAGAVAGAIINSGQDCTA
MLBr_02573|M.leprae_Br4923 GAIAGNEIKPTVLELGGSDPFIVMPSADLDAAVATAVTGRVQNNGQSCIA
* *****. . :*: .** *. . ** *
Mb0791|M.bovis_AF2122/97 TTRLVVPRARYEEAVAIAAATMSSIRPGDPNDPGTVCGPLISARQRDRVQ
Rv0768|M.tuberculosis_H37Rv TTRLVVPRARYEEAVAIAAATMSSIRPGDPNDPGTVCGPLISARQRDRVQ
TH_3080|M.thermoresistible__bu LTRILMPAGRAAEFTDALAAEMESLVIGDPTDAETQIGPLVARRQQERVR
MAV_2608|M.avium_104 GSRLLVDRSVADALVEKIVARAATIKLGDPKDPTTEMGPVSNLPQYEKVL
MMAR_3556|M.marinum_M ATRAIVARELYHDFVAGVADVMSTVVIGDPMDPDTDIGPLISLAHRDRVA
MUL_2789|M.ulcerans_Agy99 ATRAIVARELCHDFVAGVADVMSTVVIGDPMDPDTDIGPLISLAHRDRVA
Mflv_4860|M.gilvum_PYR-GCK ATRAIVAEGLYDDFVAGVAEVMSKVVIGDPADPDTDLGPLISAVHREKVA
Mvan_1572|M.vanbaalenii_PYR-1 ATRAIVARDLYDDFVAGVAEVMSNIVVGDPGDPDTDLGPLITLAHRAKVA
MSMEG_1665|M.smegmatis_MC2_155 ATRAIVARELYDDFVAGVAEVMGKIVVGDPHDPDTDLGPLISYAHRDKVA
MAB_3328|M.abscessus_ATCC_1997 ATRALVARDLYDDFVAGVGEVMSKVVVGDPLDPDTDIGPLISAAHRAKVS
MLBr_02573|M.leprae_Br4923 AKRFIAHADIYDEFVEKFVARMETLRVGDPTDPDTDVGPLATESGRAQVE
.* : . .: *** *. * **: :*
Mb0791|M.bovis_AF2122/97 GYLDLAVAEGGRFACGG-ARPADREVGFYIEPTVIAGLTNDARVAREEIF
Rv0768|M.tuberculosis_H37Rv GYLDLAVAEGGRFACGG-ARPADREVGFYIEPTVIAGLTNDARVAREEIF
TH_3080|M.thermoresistible__bu GYIEAGCAEGARLVTGGPGLPAGVDRGWYVQPTLFADADNSMRIAREEIF
MAV_2608|M.avium_104 SHFASAREQGATVAYGG--EPASDLGGFFVKPTVLTGVDPSMRAVAEEIF
MMAR_3556|M.marinum_M DIVAKAPAQGARIVVGG---RIPDLAGAFYLPTLLADVGEQSQAYREEIF
MUL_2789|M.ulcerans_Agy99 DILAKAPAQGARIVVGG---RIPDLAGAFYLPTLLADVGEQSQAYREEIF
Mflv_4860|M.gilvum_PYR-GCK GMVERAPGEGGRVVTGG---VAPDGPGSFYRPTLIADVAESSEVWRDEIF
Mvan_1572|M.vanbaalenii_PYR-1 GMVERAPGEGGRIVTGG---VAPDGPGSFYRPTLIADVSEQSEVWRDEIF
MSMEG_1665|M.smegmatis_MC2_155 GMVARAPQQGGRVVTGG---EAPDLPGAFYRPTLIADVDESSEVYRDEIF
MAB_3328|M.abscessus_ATCC_1997 SIVDRAPAQGGRIVTGA---TAPDLPGSFYRPTLIADVAETSEVYRDEIF
MLBr_02573|M.leprae_Br4923 QQVDAAAAAGAVIRCGG---KRPDRPGWFYPPTVITDITKDMALYTDEVF
. . *. . *. * : **:::. :*:*
Mb0791|M.bovis_AF2122/97 GPVLTVIAHDGDDDAVRIANDSPYGLSGTVYGADPQRAARIASRLRVGTV
Rv0768|M.tuberculosis_H37Rv GPVLTVIAHDGDDDAVRIANDSPYGLSGTVYGADPQRAARIASRLRVGTV
TH_3080|M.thermoresistible__bu GPVLTVIPYPDEDAAVAIANDSDYGLAGSVFTADTERGLAIAARIRTGTF
MAV_2608|M.avium_104 GPVLAVMTFEDEDEAVAAANGTEFGLAAAVWTKDVHRAHRVAAKLRAGTV
MMAR_3556|M.marinum_M GPVLTVRAHDGDDDALRQANDTDYGLAASAWTRDVYRAQRASREISAGCV
MUL_2789|M.ulcerans_Agy99 GPVLTVRAHDGDDDALRQANDTDYGLAASAWTRDVYRAQRASREISAGCV
Mflv_4860|M.gilvum_PYR-GCK GPVLTVRSFTDDDDAIRQANDTKYGLAASAWTRDVYRAQRASREIHAGCV
Mvan_1572|M.vanbaalenii_PYR-1 GPVLAVRSFTDDDDALRQANDTAYGLAASAWTRDVYRAQRASREIHAGCV
MSMEG_1665|M.smegmatis_MC2_155 GPVLTVRSFTDDDDALRQANDTEYGLAASAWTRDVYRAQRASREINAGCV
MAB_3328|M.abscessus_ATCC_1997 GPVLTVRPFTDDDDALRQANDTAYGLAASAWTRDVYRAQRASREIRAGCV
MLBr_02573|M.leprae_Br4923 GPVASVYCATNIDDAIEIANATPFGLGSNAWTQNESEQRHFIDNIVAGQV
*** :* . * *: ** : :**.. .: : . .: .* .
Mb0791|M.bovis_AF2122/97 NVNGGVWYCADAPFGGYKQSGIGREMGLLGFEEYLEAKLIATAAN-----
Rv0768|M.tuberculosis_H37Rv NVNGGVWYCADAPFGGYKQSGIGREMGLLGFEEYLEAKLIATAAN-----
TH_3080|M.thermoresistible__bu GVNQGYTMDPFAPFGGVKGSGYGRELGPEGIDAYTDVKSISVAPASTT--
MAV_2608|M.avium_104 WVNAYRVVAPHVPFGGIGYSGIGRENGIDAVKDFTETKAVWVELSGETRD
MMAR_3556|M.marinum_M WINDHIPIVSEMPHGGVAASGFGKDMSEYSFEEYLTIKHVMSDITGVAEK
MUL_2789|M.ulcerans_Agy99 WINDHIPIVSEMPHGGVAASGFGNDMSEYSFDEYLTIKHVMSDITGVAEK
Mflv_4860|M.gilvum_PYR-GCK WINDHIPIISEMPHGGFGASGFGKDMSQYSLEEYLSIKHVMSDITSAVDK
Mvan_1572|M.vanbaalenii_PYR-1 WINDHIPIISEMPHGGFGASGFGKDMSQYSLEEYLSIKHVMSDITGVADK
MSMEG_1665|M.smegmatis_MC2_155 WINDHIPIISEMPHGGVGASGFGKDMSDYSFEEYLTIKHVMSDITGVAEK
MAB_3328|M.abscessus_ATCC_1997 WINDHIPIISEMPHGGFGASGFGKDMSDYSLEEYLTVKHVMSDITGTADK
MLBr_02573|M.leprae_Br4923 FINGMTVSYPELPFGGIKRSGYGRELSTHGIREFCNIKTVWIA-------
:* . *.** ** *.: . .. : * :
Mb0791|M.bovis_AF2122/97 -----------
Rv0768|M.tuberculosis_H37Rv -----------
TH_3080|M.thermoresistible__bu -----------
MAV_2608|M.avium_104 PFTLG------
MMAR_3556|M.marinum_M PWHRTIFSTRI
MUL_2789|M.ulcerans_Agy99 PWHRTIFSTRI
Mflv_4860|M.gilvum_PYR-GCK DWHRTIFTKR-
Mvan_1572|M.vanbaalenii_PYR-1 TWHRTIFTKR-
MSMEG_1665|M.smegmatis_MC2_155 EWHRTIFNKR-
MAB_3328|M.abscessus_ATCC_1997 DWHRTIFAKR-
MLBr_02573|M.leprae_Br4923 -----------