For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTDHSTLIDAIPTDLWIGGKQVPSSTGTRFPVHNPATGAVLTTVADAAASDGAIALDEATAVQRSWAATA PRERAEILRRAWELVIARRDDFALAMTLEMGKPLAESQGEVAYGGEFLRWFSEEAVRINGRYTSSPTGAG RILVAKQPIGPTLAITPWNFPLAMGTRKIGPALAAGCTMIVKPAAETPLTMLLLGQVFADAGLPAGVLSI LPTTNASALTGPLIDDPRLRKLTFTGSTGVGKILLSQCATRVLRSSMELGGNAPFVVFDDADVDAAVDGA MAAKMRNIGEACTAANRFHVDNAVRAEFTEKLTARMGALTIGPGDENGVQVGPLITAKQRASVDELVQDA VSKGAVVTTGGVVADGEGYFYPPTVLTDVPADARILREEVFGPVAAITGFDGEEAGIAAANDTEFGLAAY VYTANLDRALRVSQAVESGMVGVNRGVISDVAAPFGGIKESGIGREAGSEGIEEYLETKYIALP
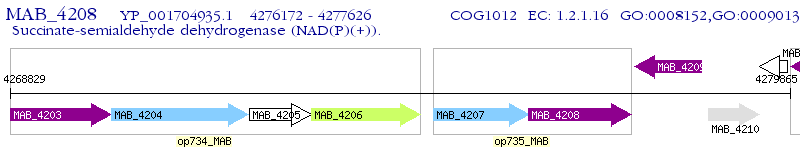
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4208 | - | - | 100% (484) | putative succinate-semialdehyde dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4418 | - | e-154 | 59.47% (454) | succinate-semialdehyde dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3328 | - | 7e-80 | 35.33% (467) | gamma-aminobutyraldehyde dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2883c | aldC | 3e-76 | 37.33% (450) | aldehyde dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1858 | - | 0.0 | 70.65% (477) | succinate-semialdehyde dehydrogenase (NAD(P)(+)) |
| M. tuberculosis H37Rv | Rv0223c | - | 1e-81 | 37.03% (478) | aldehyde dehydrogenase |
| M. leprae Br4923 | MLBr_02573 | gabD1 | 1e-69 | 34.89% (450) | succinic semialdehyde dehydrogenase |
| M. marinum M | MMAR_3556 | - | 2e-82 | 36.83% (467) | aldehyde dehydrogenase NAD dependent |
| M. avium 104 | MAV_2101 | - | 4e-82 | 36.52% (471) | aldehyde dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5538 | - | 0.0 | 69.60% (477) | [NADP+] succinate-semialdehyde dehydrogenase [Mycobacterium |
| M. thermoresistible (build 8) | TH_3381 | - | 1e-148 | 73.95% (357) | PUTATIVE Succinate-semialdehyde dehydrogenase (NAD(P)(+)) |
| M. ulcerans Agy99 | MUL_2789 | - | 3e-80 | 36.19% (467) | gamma-aminobutyraldehyde dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_4876 | - | 0.0 | 69.85% (481) | succinate-semialdehyde dehydrogenase (NAD(P)(+)) |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1858|M.gilvum_PYR-GCK MDNSAVEALLASVPTGLWIGGEEREGTS--TFDVLDPSDDTVLTSVADAT
Mvan_4876|M.vanbaalenii_PYR-1 MDTS---ALLKSVPTGLWIGGEERQGTS--TFSVLDPSDDEVLATVADAT
MSMEG_5538|M.smegmatis_MC2_155 MRIE---DLISSVPTGLWIGGEEREAAS--TFNVLDPSDDQVITAVADAT
TH_3381|M.thermoresistible__bu --------------------------------------------------
MAB_4208|M.abscessus_ATCC_1997 MTDHS--TLIDAIPTDLWIGGKQVPSSTGTRFPVHNPATGAVLTTVADAA
MMAR_3556|M.marinum_M ------MTTTVSVAGSWING--AAHSTGGRQHAVINPATQETIAELALAT
MUL_2789|M.ulcerans_Agy99 ------MTTTVSVAGSWING--AAHSTGGRQHAVINPATQETIAELALAT
Rv0223c|M.tuberculosis_H37Rv ------MSDSATEYDKLFIGGKWTKPSTSDVIEVRCPATGEYVGKVPMAA
MAV_2101|M.avium_104 ---------------------------MSTTIPVFNPSTEEQIAEVPDSD
Mb2883c|M.bovis_AF2122/97 ----------------------------MSTTQLINPATEEVLASVDHTD
MLBr_02573|M.leprae_Br4923 -----------------------------MSIATINPATGETVKTFTPTT
Mflv_1858|M.gilvum_PYR-GCK ADDARAALDAACAVQAE--WAGTAPRKRGEILRSVFETITARADDIAALM
Mvan_4876|M.vanbaalenii_PYR-1 ADDARDALDAACAVQGE--WAATPARKRGEILRSVFETITARADDIAALM
MSMEG_5538|M.smegmatis_MC2_155 AEDAIAALDAACAVQDE--WAATPARTRGEILRSVFEKITERADDIAALM
TH_3381|M.thermoresistible__bu -----------------------VVRRRG---------------------
MAB_4208|M.abscessus_ATCC_1997 ASDGAIALDEATAVQRS--WAATAPRERAEILRRAWELVIARRDDFALAM
MMAR_3556|M.marinum_M PADVDRAVVAARRALPQ--WAGVTPAERSAVLAKLADLLEQRAPELIAEE
MUL_2789|M.ulcerans_Agy99 PADVDHAVVAARRALPQ--WAGVTTAERSAVLAELADLLEQRAPELIAEE
Rv0223c|M.tuberculosis_H37Rv AADVDAAVAAARAAFDNGPWPSTPPHERAAVIAAAVKMLAERKDLFTKLL
MAV_2101|M.avium_104 QAAVDAAVARARETFESGVWRKAPASHRANVLFRAARIIEERKDELAALE
Mb2883c|M.bovis_AF2122/97 ANAVDDAVQRARAAQRR--WARLAPAQRAAGLRAFAAAVQAHLDELAALE
MLBr_02573|M.leprae_Br4923 QDEVEDAIARAYARFDN--YRHTTFTQRAKWANATADLLEAEANQSAALM
*.
Mflv_1858|M.gilvum_PYR-GCK TLEMGKVLAESKGEVTY-GAEFFRWFAEEAVRIDGRYTAAPAGT------
Mvan_4876|M.vanbaalenii_PYR-1 TLEMGKVFAESKGEVTY-GAEFFRWFAEEAVRIEGRYTPAPAGT------
MSMEG_5538|M.smegmatis_MC2_155 TLEMGKVLPESKGEVAY-GAEFFRWFSEEAVRIDGRYTHAPAGT------
TH_3381|M.thermoresistible__bu ----------------------------------GPPPRAPAGN------
MAB_4208|M.abscessus_ATCC_1997 TLEMGKPLAESQGEVAY-GGEFLRWFSEEAVRINGRYTSSPTGA------
MMAR_3556|M.marinum_M VCQTGKPLRLAEEFDVPGSVDNIRFFAGAARRLPGQATAEYSAD------
MUL_2789|M.ulcerans_Agy99 VCQTGKPLRLAEEFDVPGSVDNIRFFAGAARRLPGQATAEYSAD------
Rv0223c|M.tuberculosis_H37Rv AAETGQPPTIIETMHWMGSMGAMNYFAGAADKVTWTETRTGSYG------
MAV_2101|M.avium_104 SQDNGMNLTAAGHIIPV-SVDMLKYYAGWVGKIHGESANLVSDGLLGTWE
Mb2883c|M.bovis_AF2122/97 VANSGHPIVSAEWEAGH-VRDVLAFYAASPERLSGRQIPVAGG-------
MLBr_02573|M.leprae_Br4923 TLEMGKTLQSAKDEAMK-CAKGFRYYAEKAETLLADEPAEAAAVG-----
Mflv_1858|M.gilvum_PYR-GCK -GRILVTKQAVGPCYAITPWNFPLAMGTRKMGPAFAAGCTMIVKPAQETP
Mvan_4876|M.vanbaalenii_PYR-1 -GRILVTKQAVGPCYAITPWNFPLAMGTRKMGPAFAAGCTMIVKPAQETP
MSMEG_5538|M.smegmatis_MC2_155 -GRILVTKQAVGPCYAITPWNFPLAMGTRKMGPAFAAGCTMIVKPAQETP
TH_3381|M.thermoresistible__bu -GRILVTKQAVGPCYAITPWNFPLAMGTRKIGPALAAGCTMIVKPAQETP
MAB_4208|M.abscessus_ATCC_1997 -GRILVAKQPIGPTLAITPWNFPLAMGTRKIGPALAAGCTMIVKPAAETP
MMAR_3556|M.marinum_M -HTSSIRREAIGVVATITPWNYPLQMAVWKVMPALAAGCTVVIKPSELTP
MUL_2789|M.ulcerans_Agy99 -HTSSIRREAIGVVATITPWNYPLQMAVWKVMPALAAGCTVVIKPSELTP
Rv0223c|M.tuberculosis_H37Rv --QSIVSREPVGVVGAIVAWNVPLFLAVNKIAPALLAGCTIVLKPAAETP
MAV_2101|M.avium_104 TYHTFTQLEPVGVVGLIIPWNGPFFVAMLKVAPALAAGCSAVLKPAEETP
Mb2883c|M.bovis_AF2122/97 --VDVTFNEPMGVVGVITPWNFPMVIASWAIAPALAAGNAVLVKPAELTP
MLBr_02573|M.leprae_Br4923 ASKALARYQPLGVVLAVMPWNFPLWQTVRFAAPALMAGNVGLLKHASNVP
:.:* : .** *: **: ** ::* : .*
Mflv_1858|M.gilvum_PYR-GCK LTMLLLAKLMADAGLPKGVLSVLPTTSPGPVTETLIDDGRLRKLTFTGST
Mvan_4876|M.vanbaalenii_PYR-1 LTMLLLAKLMAEAGLPKGVLSVLPTSNPGAVTEALINDGRLRKLTFTGST
MSMEG_5538|M.smegmatis_MC2_155 LTMLLLAKLMDEAGLPKGVLSVLPTSNPRGVTGALIDDGRLRKLTFTGST
TH_3381|M.thermoresistible__bu LTMLLLAKLIDEAGLPKGVLSVLPTSNPRPLTEALIGDGRLRKLTFTGST
MAB_4208|M.abscessus_ATCC_1997 LTMLLLGQVFADAGLPAGVLSILPTTNASALTGPLIDDPRLRKLTFTGST
MMAR_3556|M.marinum_M LTTLTLARLAAEAGLPDGVLNVVTGL-GADVGAALAAHPGVDLVTFTGST
MUL_2789|M.ulcerans_Agy99 LTTLTLARLAAEAGLPDGVLNVVTGL-GADVGAALAAHPGVDLVTFTGST
Rv0223c|M.tuberculosis_H37Rv LTANALAEVFAEVGLPEGVLSVVPG--GIETGQALTSNPDIDMFTFTGSS
MAV_2101|M.avium_104 LTALKLEEIFREAGLPDGVLNVITGY-GETTGAALTAHPDVDKISFTGST
Mb2883c|M.bovis_AF2122/97 LTTMRLGELAVEAGLDEDLLQVLPGK-GTVVGERFVTHPDIRKIVFTGST
MLBr_02573|M.leprae_Br4923 QCALYLADVIARGGFPEDCFQTLLIS--ANGVEAILRDPRVAAATLTGSE
* : *: . :. : : . : :***
Mflv_1858|M.gilvum_PYR-GCK GVGKALVKQSADKLLRTSMELGGNAPFVVFDDADVDAAVDGAILAKMRNG
Mvan_4876|M.vanbaalenii_PYR-1 GVGKALVKQSADKLLRTSMELGGNAPFIVFDDADVDAAVDGAILAKMRNG
MSMEG_5538|M.smegmatis_MC2_155 GVGKALVKQSADKLLRTSMELGGNAPFIVFDDADVDAAVDGAILAKMRNG
TH_3381|M.thermoresistible__bu GVGKALAQQASAKLLRTSMELGGNAPFIVFDDADIDAAVDGAMLAKMRNG
MAB_4208|M.abscessus_ATCC_1997 GVGKILLSQCATRVLRSSMELGGNAPFVVFDDADVDAAVDGAMAAKMRNI
MMAR_3556|M.marinum_M AVGRKVMAAAATHGRRTQLELGGKAPFVVFDDADLDAAIQGAVAGALINS
MUL_2789|M.ulcerans_Agy99 AVGRKVMAAAATHGRRTQLELGGKAPFVVFDDADLDAAIQGAVAGAVINS
Rv0223c|M.tuberculosis_H37Rv AVGREVGRRAAEMLKPCTLELGGKSAAIILEDVDLAAAIPMMVFSGVMNA
MAV_2101|M.avium_104 EVGRLIVKAAAGNLKRLTLELGGKSPLIMFDDANLDKAIMGAGMGLLAGS
Mb2883c|M.bovis_AF2122/97 EVGKRVMAGAAAQVKRVTLELGGKSANIVFHDCDLERAATTAPAGVFDNA
MLBr_02573|M.leprae_Br4923 PAGQAVGAIAGNEIKPTVLELGGSDPFIVMPSADLDAAVATAVTGRVQNN
.*: : .. :****. . ::: . :: * . . .
Mflv_1858|M.gilvum_PYR-GCK GEACTAANRIHVANAVREEFTDKFVKRMSEFTLGKGLDEKSTLGPLINAK
Mvan_4876|M.vanbaalenii_PYR-1 GEACTAANRIHVANAVREEFTEKFVKRMSEFTLGKGLDEKSTLGPLINAK
MSMEG_5538|M.smegmatis_MC2_155 GEACTAANRFHVANSVREEFTEKLVKRMSEFTLGKGLDPSSTLGPLINSK
TH_3381|M.thermoresistible__bu GEACTAANRFHVANAVREEFTEKLVKRMSEVTLGNGLDGSSTLGPLINAQ
MAB_4208|M.abscessus_ATCC_1997 GEACTAANRFHVDNAVRAEFTEKLTARMGALTIGPGDENGVQVGPLITAK
MMAR_3556|M.marinum_M GQDCTAATRAIVARELYHDFVAGVADVMSTVVIGDPMDPDTDIGPLISLA
MUL_2789|M.ulcerans_Agy99 GQDCTAATRAIVARELCHDFVAGVADVMSTVVIGDPMDPDTDIGPLISLA
Rv0223c|M.tuberculosis_H37Rv GQGCVNQTRILAPRSRYDEIVAAVTNFVTALPVGPPSDPAAQIGPLISEK
MAV_2101|M.avium_104 GQNCSCTSRIYVQRGIYDQVVEGLAEFAMMLPMGGSDDPNSVLGPLISEK
Mb2883c|M.bovis_AF2122/97 GQDCCARSRILVQRSVYDRFMELLEPAVHSIVVGDPGSRATEMGPLVSRA
MLBr_02573|M.leprae_Br4923 GQSCIAAKRFIAHADIYDEFVEKFVARMETLRVGDPTDPDTDVGPLATES
*: * .* . . . . :* . :*** .
Mflv_1858|M.gilvum_PYR-GCK QVATVTELVSDAVSRGATVAVGGVAPGGPG--NFYPATVLTDVPADARIL
Mvan_4876|M.vanbaalenii_PYR-1 QVATVTELVSDAVSRGATVAVGGVAPGGPG--NFYPATVLTDVPADARIL
MSMEG_5538|M.smegmatis_MC2_155 QVATVTELVSDAVSRGATVAVGGVAPGGPG--NFYPATVLADVPADARIL
TH_3381|M.thermoresistible__bu QRDTVVELVTDAVERGATVAVGGTVPDGPG--WFYPATVLTDVSADARIL
MAB_4208|M.abscessus_ATCC_1997 QRASVDELVQDAVSKGAVVTTGGVVADGEG--YFYPPTVLTDVPADARIL
MMAR_3556|M.marinum_M HRDRVADIVAKAPAQGARIVVGGRIPDLAG--AFYLPTLLADVGEQSQAY
MUL_2789|M.ulcerans_Agy99 HRDRVADILAKAPAQGARIVVGGRIPDLAG--AFYLPTLLADVGEQSQAY
Rv0223c|M.tuberculosis_H37Rv QRTRVEGYIAKGIEEGARLVCGGGRPEGLDNGFFIQPTVFADVDNKMTIA
MAV_2101|M.avium_104 QRQRVEGIVNDGVAAGAKVITGG-KPMDRK-GYFYEATIITNTTPDMRLI
Mb2883c|M.bovis_AF2122/97 HRDKVAGYVPD----DAPVAFRGTAPAGRG--FWFPPTVLTPKRGDRTVT
MLBr_02573|M.leprae_Br4923 GRAQVEQQVDAAAAAGAVIRCGGKRPDRPG--WFYPPTVITDITKDMALY
* : .* : * . : .*::: .
Mflv_1858|M.gilvum_PYR-GCK KEEVFGPVAPIAGFDTEEEGIAAANDTEYGLAAYVYTQSLDRALRVAEGI
Mvan_4876|M.vanbaalenii_PYR-1 KEEVFGPVAPIAGFDTEEEGIAAANDTEYGLAAYVYTQSLDRALRVAENI
MSMEG_5538|M.smegmatis_MC2_155 KEEVFGPVAPITGFDTEEEGIAAANDTEYGLAAYIYTQSLDRALRVAETI
TH_3381|M.thermoresistible__bu KEEVFGPVAPIVGFDTEEEGIAAANDTEYGLAAYIYTRSLDRALRVAEAI
MAB_4208|M.abscessus_ATCC_1997 REEVFGPVAAITGFDGEEAGIAAANDTEFGLAAYVYTANLDRALRVSQAV
MMAR_3556|M.marinum_M REEIFGPVLTVRAHDGDDDALRQANDTDYGLAASAWTRDVYRAQRASREI
MUL_2789|M.ulcerans_Agy99 REEIFGPVLTVRAHDGDDDALRQANDTDYGLAASAWTRDVYRAQRASREI
Rv0223c|M.tuberculosis_H37Rv QEEIFGPVLAIIPYDTEEDAIAIANDSVYGLAGSVWTTDVPKGIKISQQI
MAV_2101|M.avium_104 REEIFGPVGCVIPFDDEDEVIAAANDTHYGLAGAIWTENLSRAHRVVNEL
Mb2883c|M.bovis_AF2122/97 -DEIFGPVVVVLTFDDEADAISLANDTAYGLSGSIWTDDLSRALRVARAV
MLBr_02573|M.leprae_Br4923 TDEVFGPVASVYCATNIDDAIEIANATPFGLGSNAWTQNESEQRHFIDNI
:*:**** : : ** : :**.. :* . . : :
Mflv_1858|M.gilvum_PYR-GCK ESGMVGVNRGVISDPAAPFGGIKESGFGREGGTEGIEEYLDTKYIALTK-
Mvan_4876|M.vanbaalenii_PYR-1 ESGMVGVNRGVISDAAAPFGGIKESGFGREGGTEGIEEYLDTKYIALTR-
MSMEG_5538|M.smegmatis_MC2_155 QSGMVGINRGVISDPAAPFGGIKESGFGREGGIEGIEEYLDTKYIALTK-
TH_3381|M.thermoresistible__bu ESGMVGVNRGVISDAAAPFGGIKESGFGREGGTEGIEEYLETKYIALT--
MAB_4208|M.abscessus_ATCC_1997 ESGMVGVNRGVISDVAAPFGGIKESGIGREAGSEGIEEYLETKYIALP--
MMAR_3556|M.marinum_M SAGCVWINDHIPIVSEMPHGGVAASGFGKDMSEYSFEEYLTIKHVMSDIT
MUL_2789|M.ulcerans_Agy99 SAGCVWINDHIPIVSEMPHGGVAASGFGNDMSEYSFDEYLTIKHVMSDIT
Rv0223c|M.tuberculosis_H37Rv RTGTYGIN-WYAFDPGSPFGGYKNSGIGRENGPEGVEHFTQQKSVLLPMG
MAV_2101|M.avium_104 RAGQIWVNSALAADPSMPISGHKQSGWGGERGRKGIEAYFNTKAVYIGL-
Mb2883c|M.bovis_AF2122/97 ESGNLSVNSHSSVRFNTPFGGFKQSGVGRELGPDAPLQFTETKNVFIAVG
MLBr_02573|M.leprae_Br4923 VAGQVFINGMTVSYPELPFGGIKRSGYGRELSTHGIREFCNIKTVWIA--
:* :* * .* ** * : . . : * :
Mflv_1858|M.gilvum_PYR-GCK ----------------
Mvan_4876|M.vanbaalenii_PYR-1 ----------------
MSMEG_5538|M.smegmatis_MC2_155 ----------------
TH_3381|M.thermoresistible__bu ----------------
MAB_4208|M.abscessus_ATCC_1997 ----------------
MMAR_3556|M.marinum_M GVAEKPWHRTIFSTRI
MUL_2789|M.ulcerans_Agy99 GVAEKPWHRTIFSTRI
Rv0223c|M.tuberculosis_H37Rv YTVA------------
MAV_2101|M.avium_104 ----------------
Mb2883c|M.bovis_AF2122/97 EEM-------------
MLBr_02573|M.leprae_Br4923 ----------------