For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VRFTGKHAIITGGGSGIGAALCRALDRAGAQVVCTDIDEAAAGAVVSTLSPRARAARLDVTDAAAVQGVV DDVVARTGRLDLIFNNAGIVWGGDTELLTLDQWNAIIDINLRGVVHGVAAAYPQMLRQGHGHIVNTASMA GLTAAGQVTSYVATKHAVVGLSMALRSEAVPRGVGVLVVCPAAVETPILDKGAVGGFVGRDYFLQTQGGR AYDADRLARDTLRAVERNKAILVKPTVAQAQWWFARLAPGLMNRMAMRFVTGQRTRQVDRYPSSAADTG
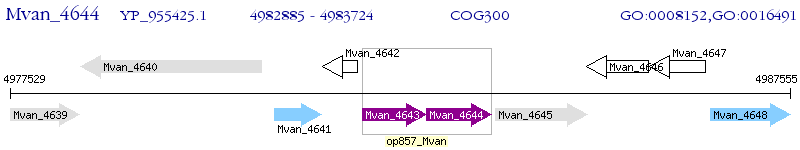
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4644 | - | - | 100% (279) | short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_3101 | - | 4e-59 | 49.45% (273) | short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_0852 | - | 3e-36 | 39.92% (243) | short-chain dehydrogenase/reductase SDR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0874c | - | 5e-31 | 39.27% (191) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_2070 | - | 1e-117 | 78.28% (267) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv0851c | - | 3e-31 | 39.27% (191) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_00862 | ephD | 1e-30 | 33.20% (259) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0090c | - | 3e-34 | 35.32% (252) | oxidoreductase EphD |
| M. marinum M | MMAR_2600 | - | 4e-35 | 35.69% (255) | short-chain dehydrogenase |
| M. avium 104 | MAV_0895 | - | 4e-32 | 34.66% (251) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5235 | - | 1e-112 | 76.21% (269) | short chain dehydrogenase family protein |
| M. thermoresistible (build 8) | TH_2445 | - | 1e-111 | 75.46% (273) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_3160 | - | 4e-35 | 35.69% (255) | short-chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5235|M.smegmatis_MC2_155 --------------------------------------------------
TH_2445|M.thermoresistible__bu --------------------------------------------------
Mvan_4644|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2070|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_00862|M.leprae_Br4923 MPATQSIHKVPHQFVESPDGVRLAIYQSGQPEGPAIVLVHGFPDSHVLWD
MAB_0090c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_2600|M.marinum_M --------------------------------------------------
MUL_3160|M.ulcerans_Agy99 --------------------------------------------------
Mb0874c|M.bovis_AF2122/97 --------------------------------------------------
Rv0851c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0895|M.avium_104 --------------------------------------------------
MSMEG_5235|M.smegmatis_MC2_155 --------------------------------------------------
TH_2445|M.thermoresistible__bu --------------------------------------------------
Mvan_4644|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2070|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_00862|M.leprae_Br4923 GVVPLLAKRFRIVRYDNRGVGQSSVPKPVSAYTMTRFADDFAAVIDELSP
MAB_0090c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_2600|M.marinum_M --------------------------------------------------
MUL_3160|M.ulcerans_Agy99 --------------------------------------------------
Mb0874c|M.bovis_AF2122/97 --------------------------------------------------
Rv0851c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0895|M.avium_104 --------------------------------------------------
MSMEG_5235|M.smegmatis_MC2_155 --------------------------------------------------
TH_2445|M.thermoresistible__bu --------------------------------------------------
Mvan_4644|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2070|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DRPVHVLAHDWGSVGVWEYLSRSGASDRVASFTSVSGPSQDQLVDYILSG
MAB_0090c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_2600|M.marinum_M --------------------------------------------------
MUL_3160|M.ulcerans_Agy99 --------------------------------------------------
Mb0874c|M.bovis_AF2122/97 --------------------------------------------------
Rv0851c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0895|M.avium_104 --------------------------------------------------
MSMEG_5235|M.smegmatis_MC2_155 --------------------------------------------------
TH_2445|M.thermoresistible__bu --------------------------------------------------
Mvan_4644|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2070|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LRRPWRPRTFARAVSQLFRLTYMVLFSIPVLVPMVLWIALSSATIRRTMV
MAB_0090c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_2600|M.marinum_M --------------------------------------------------
MUL_3160|M.ulcerans_Agy99 --------------------------------------------------
Mb0874c|M.bovis_AF2122/97 --------------------------------------------------
Rv0851c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0895|M.avium_104 --------------------------------------------------
MSMEG_5235|M.smegmatis_MC2_155 --------------------------------------------------
TH_2445|M.thermoresistible__bu --------------------------------------------------
Mvan_4644|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2070|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DNISTDQIHHSATLARDAAHSVKTYPANYFRAFSGRRRGKGVPVVNVPVQ
MAB_0090c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_2600|M.marinum_M --------------------------------------------------
MUL_3160|M.ulcerans_Agy99 --------------------------------------------------
Mb0874c|M.bovis_AF2122/97 --------------------------------------------------
Rv0851c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0895|M.avium_104 --------------------------------------------------
MSMEG_5235|M.smegmatis_MC2_155 --------------------------------------------------
TH_2445|M.thermoresistible__bu --------------------------------------------------
Mvan_4644|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2070|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LIVNTMDRYVRPYGYDATARWVPRLWRRDIKAGHFSPMSHPQVMAAAVHD
MAB_0090c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_2600|M.marinum_M --------------------------------------------------
MUL_3160|M.ulcerans_Agy99 --------------------------------------------------
Mb0874c|M.bovis_AF2122/97 --------------------------------------------------
Rv0851c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0895|M.avium_104 --------------------------------------------------
MSMEG_5235|M.smegmatis_MC2_155 ---MS--------------------FTGKTAIVTGAGSGIGATLCRALVD
TH_2445|M.thermoresistible__bu ---MT--------------------FAGKHALVTGAGSGIGAALCRALAA
Mvan_4644|M.vanbaalenii_PYR-1 ---MR--------------------FTGKHAIITGGGSGIGAALCRALDR
Mflv_2070|M.gilvum_PYR-GCK ---MSSTAD----------------FAGKQALLTGGGSGIGAALCRALDR
MLBr_00862|M.leprae_Br4923 LADLVDGKQPSRALLRAQVGRSRDTFGDTLVSVTGAGSGIGRETALALAR
MAB_0090c|M.abscessus_ATCC_199 ---MTSG--------SAQR--------NSLVVVTGAGSGIGRETSLAFAR
MMAR_2600|M.marinum_M MFKLNSGTN------TSDLAG-------RVVAITGAGSGIGRELALLCAQ
MUL_3160|M.ulcerans_Agy99 MFKLNSGTN------TSDLAG-------RVVAITGAGSGIGRELALLCAQ
Mb0874c|M.bovis_AF2122/97 ---MDGFP-------------------GRGAVITGGASGIGLATGTEFAR
Rv0851c|M.tuberculosis_H37Rv ---MDGFP-------------------GRGAVITGGASGIGLATGTEFAR
MAV_0895|M.avium_104 ---MDGFLS---------------GFDGRAAVVTGGASGIGLATATEFAR
: . :**..****
MSMEG_5235|M.smegmatis_MC2_155 AGADVLCTDVDADAAARTAAARTAAALGARSARLDVTDAAAVQTTVDDVV
TH_2445|M.thermoresistible__bu AGAEVMCTDIDG-----AAAERTAQSIGGRWAALDVTDAAAVQAAVDGVV
Mvan_4644|M.vanbaalenii_PYR-1 AGAQVVCTDIDEAA---AGAVVSTLSPRARAARLDVTDAAAVQGVVDDVV
Mflv_2070|M.gilvum_PYR-GCK AGAHVVCTDIDELA---AERVVSTLSSRARAARLDVTDSAAVQAAVDDVE
MLBr_00862|M.leprae_Br4923 RGAEVVVSDIDEAAAKNTAAQIVASGGVAYAYALDVSDAAAVEAFAEQVS
MAB_0090c|M.abscessus_ATCC_199 QGARVVVADINHDTANETVGLIEKLGGSAYPYALDVSDEAAVTAFADDVC
MMAR_2600|M.marinum_M RGADLALCDINDTAVADTAQTARGFGHDVITRRVDVSDPEQMTAFADATL
MUL_3160|M.ulcerans_Agy99 RGADLALCDINDTAVADTAQTARGFGHDVITRRVDVSDPEQMTAFADATL
Mb0874c|M.bovis_AF2122/97 RGARVVLGDVDKPGLRQAVNHLRAEGFDVHGVMCDVRHREEVTHLADEAF
Rv0851c|M.tuberculosis_H37Rv RGARVVLGDVDKPGLRQAVNHLRAEGFDVHSVMCDVRHREEVTHLADEAF
MAV_0895|M.avium_104 RGARLVLSDVDQPALEQAVNGLRGQGFDAHGVVCDVRHLDEMVRLADEAF
** : *:: : . ** . : .: .
MSMEG_5235|M.smegmatis_MC2_155 ARAGRLDLMFNNAGIVWGGDTELLTLDQWNAIIDVNIRGVVHGVAAAYPQ
TH_2445|M.thermoresistible__bu DRAGRLDLLFNNAGIVWGGDTELLTLDQWNAIIDVNIRGVVHGVAAAYPL
Mvan_4644|M.vanbaalenii_PYR-1 ARTGRLDLIFNNAGIVWGGDTELLTLDQWNAIIDINLRGVVHGVAAAYPQ
Mflv_2070|M.gilvum_PYR-GCK ARAGRLDLMFNNAGIVFGGETELLTLDQWNAIIDVNIRGVVHGVAAAYPL
MLBr_00862|M.leprae_Br4923 AKHGVPDIVVNNAGIGQAGRFLDTPPEEFDRVLAVNLGGVVNCCRAFGQR
MAB_0090c|M.abscessus_ATCC_199 AKHGVPDVLINNAGVGHGGRFFETSSTDFQRVLNINLGGVVHGCRAFGPR
MMAR_2600|M.marinum_M GHFGGVDLLVNNAGVGLIGGFLDTSRKDWDWLVSINVMGVVHGCEAFLPA
MUL_3160|M.ulcerans_Agy99 GHFGGVDLLVNNAGVGLIGGFLDTSRKDWDWLVSINVMGVVHGCEAFLPA
Mb0874c|M.bovis_AF2122/97 RLLGHFDVVFSNAGIVVGGPIVEMTHDDWRWVIDVDLWGSIHTVEAFLPR
Rv0851c|M.tuberculosis_H37Rv RLLGHVDVVFSNAGIVVGGPIVEMTHDDWRWVIDVDLWGSIHTVEAFLPR
MAV_0895|M.avium_104 RLLGGVDVVFSNAGIVVAGPLAQMNHDDWRWVIDIDLWGSIHAVEAFLPR
* *::..***: * :: :: ::: * :: *
MSMEG_5235|M.smegmatis_MC2_155 MIRQGHG-HIVNTASMAGLAAAGQLTSYVMSKHAVVGLSLALRSEAAAHG
TH_2445|M.thermoresistible__bu MVRQGHG-HIVNTASMAGLAAAGQLTSYVMTKHAVVGLSLALRSEAAAHG
Mvan_4644|M.vanbaalenii_PYR-1 MLRQGHG-HIVNTASMAGLTAAGQVTSYVATKHAVVGLSMALRSEAVPRG
Mflv_2070|M.gilvum_PYR-GCK MLRQGHG-HIVNTASMAGLAAAGHLTSYVATKHAVVGLSLALRSEALSRG
MLBr_00862|M.leprae_Br4923 LVERGTGGHIVNVSSMAAYAPLQSLSAYCTSKAATYMFSDCLRAELDAVG
MAB_0090c|M.abscessus_ATCC_199 MAERKSG-HIVNISSAAAYTPIPEMGAYATSKAAVFMFSDVLRGELARDK
MMAR_2600|M.marinum_M MIESGRGGHVVNLSSAAGLLANSALSAYSATKFAVLGLSEALRIELEPHR
MUL_3160|M.ulcerans_Agy99 MIESGRGGHVVNLSSAAGLLANPALSAYSATKFAVLGLSEALRIELEPHR
Mb0874c|M.bovis_AF2122/97 LLEQGTGGHVVFTASFAGLVPNAGLGAYGVAKYGVVGLAETLAREVTADG
Rv0851c|M.tuberculosis_H37Rv LLEQGTGGHVVFTASFAGLVPNAGLGAYGVAKYGVVGLAETLAREVTADG
MAV_0895|M.avium_104 LLEQGTGGHIAFTASFAGLVPNAGLGTYGVAKYGVVGLAETLAREVKPNG
: . * *:. :* *. . : :* :* .. :: * *
MSMEG_5235|M.smegmatis_MC2_155 VGVLAVCPAAVETPILDKGAVGGFVGRDYFLRGQGMKT-------AYDPD
TH_2445|M.thermoresistible__bu VGVLAVCPAAVETPLLDKGAVGGFLGRDYFIRGQGVRS-------AYDPD
Mvan_4644|M.vanbaalenii_PYR-1 VGVLVVCPAAVETPILDKGAVGGFVGRDYFLQTQGGR--------AYDAD
Mflv_2070|M.gilvum_PYR-GCK IGVLAVCPAAVETPILDKGAVGSFGGRDFFLRAQGGK--------AYDAD
MLBr_00862|M.leprae_Br4923 VGLTTICPGVIDTNIIQSTRIDTPTAKQERVRDRRGQLDMMFRLRRYGPD
MAB_0090c|M.abscessus_ATCC_199 VKVSTICPGIVNTNIIRTTQFSGLSPEDEAKRQQQG--AGLYAKRGYGPE
MMAR_2600|M.marinum_M IGVTAICPGVINTAITKASPIRGAG-DTEGRRQRLT---VTYQRRGYTPA
MUL_3160|M.ulcerans_Agy99 IGVTAICPGVINTAITKASPIRGAG-DTEGRRQRLT---VTYQRRGYTPA
Mb0874c|M.bovis_AF2122/97 IGVSVLCPMVVETNLVANSERIRGAACAQSST-TGSPGPLPLQDDNLGVD
Rv0851c|M.tuberculosis_H37Rv IGVSVLCPMVVETNLVANSERIRGAACAQSST-TGSPGPLPLQDDNLGVD
MAV_0895|M.avium_104 IGVSVLCPMVVETKLVSNSERIRGADYGMSATPEGAFGPLPTQDESVSAD
: : .:** ::* :
MSMEG_5235|M.smegmatis_MC2_155 RLAADTLRAIERNKALLVKPRRAHASWLLARLAPGLMQRLSVRFVAAQRA
TH_2445|M.thermoresistible__bu RLASDTLRAVQRNKALLVKPRVAHAGWLFARLAPGLMQRASIRFVERQRA
Mvan_4644|M.vanbaalenii_PYR-1 RLARDTLRAVERNKAILVKPTVAQAQWWFARLAPGLMNRMAMRFVTGQRT
Mflv_2070|M.gilvum_PYR-GCK RLAREVLDAITRNKAVLVKPRVAHAQWLLNRLAPTGLNRLASRFLAAQQS
MLBr_00862|M.leprae_Br4923 KVADTILSAIKKNKPIRPVAPEAYALYGISRVLPQVLRSTARIRVI----
MAB_0090c|M.abscessus_ATCC_199 KVAKEIVNAVAKGKSVVPVTPEAHVQYHFSRLAPALNRFFWGLSDKISR-
MMAR_2600|M.marinum_M RAARNILRAVERNKAVAPVAAEAHVMYVLSRIMPPLARWVSAQMAKAAK-
MUL_3160|M.ulcerans_Agy99 RAARNILRAVERNKAVAPVAAEAHVMYVLSRIMPPLARWVSAQMAKAAK-
Mb0874c|M.bovis_AF2122/97 DIAQLTADAILANR--LYVLPHAASRASIRRRFERIDRTFDEQAAEGWRH
Rv0851c|M.tuberculosis_H37Rv DIAQLTADAILANR--LYVLPHAASRASIRRRFERIDRTFDEQAAEGWRH
MAV_0895|M.avium_104 DVARLTADAILANR--LYILPHAAARESIRRRFERIDRTFDEQAAEGWTH
* *: .: * : * .
MSMEG_5235|M.smegmatis_MC2_155 SQARTANH------
TH_2445|M.thermoresistible__bu GQRAAQVGR-----
Mvan_4644|M.vanbaalenii_PYR-1 RQVDRYPSSAADTG
Mflv_2070|M.gilvum_PYR-GCK SRVNSG--------
MLBr_00862|M.leprae_Br4923 --------------
MAB_0090c|M.abscessus_ATCC_199 --------------
MMAR_2600|M.marinum_M --------------
MUL_3160|M.ulcerans_Agy99 --------------
Mb0874c|M.bovis_AF2122/97 --------------
Rv0851c|M.tuberculosis_H37Rv --------------
MAV_0895|M.avium_104 --------------