For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DGFLSGFDGRAAVVTGGASGIGLATATEFARRGARLVLSDVDQPALEQAVNGLRGQGFDAHGVVCDVRHL DEMVRLADEAFRLLGGVDVVFSNAGIVVAGPLAQMNHDDWRWVIDIDLWGSIHAVEAFLPRLLEQGTGGH IAFTASFAGLVPNAGLGTYGVAKYGVVGLAETLAREVKPNGIGVSVLCPMVVETKLVSNSERIRGADYGM SATPEGAFGPLPTQDESVSADDVARLTADAILANRLYILPHAAARESIRRRFERIDRTFDEQAAEGWTH*
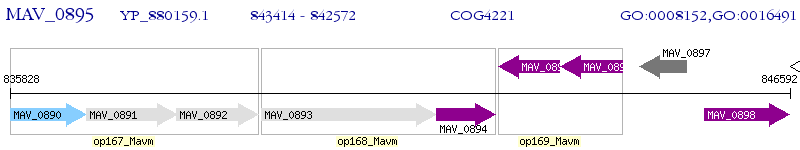
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0895 | - | - | 100% (280) | short chain dehydrogenase |
| M. avium 104 | MAV_3924 | - | 6e-31 | 38.42% (203) | short chain dehydrogenase |
| M. avium 104 | MAV_1797 | - | 3e-30 | 33.33% (252) | oxidoreductase, short chain dehydrogenase/reductase family |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0874c | - | 1e-125 | 78.62% (276) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4800 | - | 1e-34 | 35.25% (261) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv0851c | - | 1e-125 | 78.62% (276) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01740 | - | 1e-29 | 38.12% (202) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4478c | - | 2e-35 | 40.32% (253) | putative short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_4755 | - | 1e-123 | 76.98% (278) | short-chain type dehydrogenase/reductase |
| M. smegmatis MC2 155 | MSMEG_2291 | - | 4e-32 | 39.02% (205) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_3454 | - | 5e-31 | 34.22% (263) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_0326 | - | 1e-120 | 75.90% (278) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0852 | - | 1e-34 | 36.15% (260) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0874c|M.bovis_AF2122/97 ------MDGFP----GRGAVITGGASGIGLATGTEFARRGARVVLGDVDK
Rv0851c|M.tuberculosis_H37Rv ------MDGFP----GRGAVITGGASGIGLATGTEFARRGARVVLGDVDK
MAV_0895|M.avium_104 ------MDGFLSGFDGRAAVVTGGASGIGLATATEFARRGARLVLSDVDQ
MMAR_4755|M.marinum_M ------MDGFA----GRGAVITGGASGIGLATATEFARRGAKVVLADIDK
MUL_0326|M.ulcerans_Agy99 ------MDGFA----GSGAVITGGASGIGLATATEFARRGAKVVLADIDK
Mflv_4800|M.gilvum_PYR-GCK ---MELAG--------RTAIVTGAASGIGAALATELARRGVSVVIGDLDD
TH_3454|M.thermoresistible__bu ---MRIDN--------SVFVVTGAGSGIGRALALSFADAGARVVAGDIDE
MAB_4478c|M.abscessus_ATCC_199 MVSLLGFGRGERRTRKVDAVVTGAASGIGRAFAVELARRGGRVVCADVDL
MLBr_01740|M.leprae_Br4923 ------MVKTAQYFVGKRCFVTGAASGIGRATALRLAAYGAELYLTDRHC
MSMEG_2291|M.smegmatis_MC2_155 ------MAQGSD-FAGKRCFVTGAASGIGRATALALAAAGAELYLTDRDA
Mvan_0852|M.vanbaalenii_PYR-1 --------MVSNHFRGKVCVVTGAASGIGLAVGAALLDHGATVVLADFDA
.:**..**** * . : * : * .
Mb0874c|M.bovis_AF2122/97 PGLRQAVNH-LRAEGFDVHGVMCDVRHREEVTHLADEAFRLLG-HFDVVF
Rv0851c|M.tuberculosis_H37Rv PGLRQAVNH-LRAEGFDVHSVMCDVRHREEVTHLADEAFRLLG-HVDVVF
MAV_0895|M.avium_104 PALEQAVNG-LRGQGFDAHGVVCDVRHLDEMVRLADEAFRLLG-GVDVVF
MMAR_4755|M.marinum_M PGLERSVEH-LRGKGFDAHGVMCDVRHLSEVNHLAAESARLLG-QIDVVF
MUL_0326|M.ulcerans_Agy99 PGLERSVEH-LRGKGFDAHGVMCDVRHLSEVNHLAAESARLLG-QIDVVF
Mflv_4800|M.gilvum_PYR-GCK DGAHATASA-IRADGGAAAALRADATSVADIEALITLASREFT-PVDIYV
TH_3454|M.thermoresistible__bu SAADTTAAT-IRDSGGHAVPIRADATTPAGIDAMISLAQETFG-PVDGYV
MAB_4478c|M.abscessus_ATCC_199 PGAKETAAL-IGER---AIEAACDVSVFDEVEALADTAEEWFGKPSDLVI
MLBr_01740|M.leprae_Br4923 DGLAQTVSEARALGAQVPEYRALDISDYDEVAAFGADIHARHP-SMDIVM
MSMEG_2291|M.smegmatis_MC2_155 DGLVQTVADARALGAEVPAHRALDISDYDQVREFAKDIHAAHG-SMDVVM
Mvan_0852|M.vanbaalenii_PYR-1 DRLT-TVAEGLCMHDGRVHSAVVDVTVQEAVQALVEGAARAHG-SLDFLF
:. * : : * .
Mb0874c|M.bovis_AF2122/97 SNAGIVVG-GPIVEMTHDDWRWVIDVDLWGSIHTVEAFLPRLLEQGTGGH
Rv0851c|M.tuberculosis_H37Rv SNAGIVVG-GPIVEMTHDDWRWVIDVDLWGSIHTVEAFLPRLLEQGTGGH
MAV_0895|M.avium_104 SNAGIVVA-GPLAQMNHDDWRWVIDIDLWGSIHAVEAFLPRLLEQGTGGH
MMAR_4755|M.marinum_M SNAGIVVA-GPIADMTHDDWRWVIDIDLWGSIHTVEAFLPKLLEHG--GH
MUL_0326|M.ulcerans_Agy99 SNAGIVVA-GPIADMTHDVWRWVIDIDLWGSIHTVEAFLPKLLEHG--GH
Mflv_4800|M.gilvum_PYR-GCK ANAGVIGK-PGLG--TESDWDVILDVNLRAHVRAARLLVPHWQSVG-SGY
TH_3454|M.thermoresistible__bu ANAGVIGP-PGLG--TDTGWDRVLDVNLRAHVRAADALVPQWISRG-TGY
MAB_4478c|M.abscessus_ATCC_199 NNAGIGAGGQVVGAAPLADWQKTLGVNLWGVIHGSHVFAPRLRALG-RGG
MLBr_01740|M.leprae_Br4923 NIAGVSVW-GTVDRLTHDQWSNVVAINLMGPIHIIETFVPAILAAGRGGH
MSMEG_2291|M.smegmatis_MC2_155 NIAGVSAW-GTVDRLSHQQWRSMVDINLMGPIHVIEEFIPPMIEARRGGH
Mvan_0852|M.vanbaalenii_PYR-1 NNAGVGGT-MPIGVATLEQWRRIVDLNLWGVIYGVHAAVPIMVRQG-SGH
**: : * : ::* . : * *
Mb0874c|M.bovis_AF2122/97 VVFTASFAGLVPNAGLGAYGVAKYGVVGLAETLAREVTADGIGVSVLCPM
Rv0851c|M.tuberculosis_H37Rv VVFTASFAGLVPNAGLGAYGVAKYGVVGLAETLAREVTADGIGVSVLCPM
MAV_0895|M.avium_104 IAFTASFAGLVPNAGLGTYGVAKYGVVGLAETLAREVKPNGIGVSVLCPM
MMAR_4755|M.marinum_M IAFTASFAGLVPNAGLGAYGVAKYGVVGLAETLSREMKDRGVGVTVLCPM
MUL_0326|M.ulcerans_Agy99 IAFTASFAGLVPNAGLGAYGVAKYGVVGLAETLSREMKDRGVGVTVLCPM
Mflv_4800|M.gilvum_PYR-GCK FVSVASAAGLLTQLGAAGYAVSKHAAVGFAEWLAVTYGDDGIGVSCVCPL
TH_3454|M.thermoresistible__bu FVSVASAAGLLTQLGAAAYAVSKHAAVGFAEWLAITYGDRGIGVSCVCPM
MAB_4478c|M.abscessus_ATCC_199 IINVASAASFAAAPRMGAYNVSKAGVLALSETLAAELSGTGVAVTVLCPT
MLBr_01740|M.leprae_Br4923 LVNVSSAAGLVALPWHAAYSASKYGLRGLSEVLRFDLARYRIGVSVVVPG
MSMEG_2291|M.smegmatis_MC2_155 LVNVSSAAGLVALPWHGAYSASKFGLRGVSEVLRFDLARHRIGVSVVVPG
Mvan_0852|M.vanbaalenii_PYR-1 IVNTASLAGLVPFPYQSLYCTTKYGVVGLSESLRYELAADGVRVSVVCPG
. .:* *.: . . * .:* . ..:* * : *: : *
Mb0874c|M.bovis_AF2122/97 VVETNLVANSERIRGAACAQSST---TGSPGPLPLQDDNLGVDDIAQLTA
Rv0851c|M.tuberculosis_H37Rv VVETNLVANSERIRGAACAQSST---TGSPGPLPLQDDNLGVDDIAQLTA
MAV_0895|M.avium_104 VVETKLVSNSERIRGADYGMSATP--EGAFGPLPTQDESVSADDVARLTA
MMAR_4755|M.marinum_M VVETRLVSNSERIRGADYGLSSAPQVTGDMGSLPAQDDSLSVDELARLTA
MUL_0326|M.ulcerans_Agy99 VVETQLVSNSERIRGADYGLSSAPQVTGDMGSLPAQDDSLSVDELARLTA
Mflv_4800|M.gilvum_PYR-GCK GVKTPLLQGIRGIDDPDARLGAS--------SIEQSAGVISAADVATVTV
TH_3454|M.thermoresistible__bu GVRTPLLGAVRDSTDTTAQLAAR--------SITSAGAVLDAAEVADLTV
MAB_4478c|M.abscessus_ATCC_199 FVKTSIVDSG-IIEESAARLAAT----------AMKWTGVSADSVARGCL
MLBr_01740|M.leprae_Br4923 AVRTPLVDTIEIAGVDRQDPRFSR------WVDRFRGHAVSAERAAEKIL
MSMEG_2291|M.smegmatis_MC2_155 AVKTGLVQTVEIAGVDREDPDVKK------WVDRFAGHAISPEKAAAKIL
Mvan_0852|M.vanbaalenii_PYR-1 NVVSRIFGTPILGRPVDAAP---------------PEDSIPAEEAARLIL
* : :. : *
Mb0874c|M.bovis_AF2122/97 DAILANRLYVLPHAASRASIRRRFERIDRTFDEQAAEGWRH---------
Rv0851c|M.tuberculosis_H37Rv DAILANRLYVLPHAASRASIRRRFERIDRTFDEQAAEGWRH---------
MAV_0895|M.avium_104 DAILANRLYILPHAAARESIRRRFERIDRTFDEQAAEGWTH---------
MMAR_4755|M.marinum_M DAILANRLYVLPHEGARTSIQRRFARIDRTFDEQAAEGWRH---------
MUL_0326|M.ulcerans_Agy99 DAILANGLYVLPHEGARTSIQRRFARIDRTFDEQAAEGWRH---------
Mflv_4800|M.gilvum_PYR-GCK EAIREERFLVLPHAELLEMYRRKGGDYDRWISGMRRYQRTLRSL------
TH_3454|M.thermoresistible__bu AAVREGRFLVLPHPEVLDMYRQKGADYDRWIAGMRRYQRSLAAER-----
MAB_4478c|M.abscessus_ATCC_199 DAHDRGQLYVLPQFDARFMWQAKRLAPDTYTKALGIAERFSRQRGN----
MLBr_01740|M.leprae_Br4923 AGVAKNRYLIYTSHDIRVLYGFKRLAWWPYGVVMKQVNVVFTRALRPSPA
MSMEG_2291|M.smegmatis_MC2_155 AGVRRNRFLIYTSADIRALYTFKRVAWWPYSVAMRQVNVLFTRALRSG--
Mvan_0852|M.vanbaalenii_PYR-1 AGVAAGEGIIAFPEKPRELWRSYAESPDSVEGYLKEVARQRRAAFATG--
. :
Mb0874c|M.bovis_AF2122/97 --------------------------
Rv0851c|M.tuberculosis_H37Rv --------------------------
MAV_0895|M.avium_104 --------------------------
MMAR_4755|M.marinum_M --------------------------
MUL_0326|M.ulcerans_Agy99 --------------------------
Mflv_4800|M.gilvum_PYR-GCK --------------------------
TH_3454|M.thermoresistible__bu --------------------------
MAB_4478c|M.abscessus_ATCC_199 --------------------------
MLBr_01740|M.leprae_Br4923 TMRCVEPIKVGVYSQPRSGEGGKSGN
MSMEG_2291|M.smegmatis_MC2_155 -------------TSPR---------
Mvan_0852|M.vanbaalenii_PYR-1 -------DAQALFRATQSAGGT----