For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MVSNHFRGKVCVVTGAASGIGLAVGAALLDHGATVVLADFDADRLTTVAEGLCMHDGRVHSAVVDVTVQE AVQALVEGAARAHGSLDFLFNNAGVGGTMPIGVATLEQWRRIVDLNLWGVIYGVHAAVPIMVRQGSGHIV NTASLAGLVPFPYQSLYCTTKYGVVGLSESLRYELAADGVRVSVVCPGNVVSRIFGTPILGRPVDAAPPE DSIPAEEAARLILAGVAAGEGIIAFPEKPRELWRSYAESPDSVEGYLKEVARQRRAAFATGDAQALFRAT QSAGGT
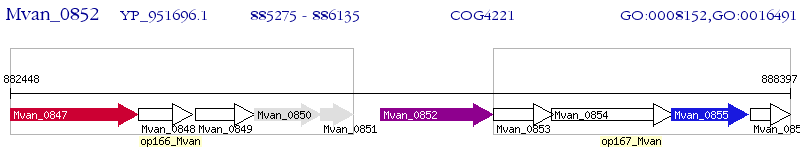
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0852 | - | - | 100% (286) | short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_3101 | - | 1e-42 | 41.67% (252) | short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_4644 | - | 3e-36 | 39.92% (243) | short-chain dehydrogenase/reductase SDR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0874c | - | 2e-35 | 44.81% (183) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_3370 | - | 2e-42 | 43.25% (252) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv0851c | - | 5e-36 | 45.36% (183) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01740 | - | 2e-31 | 37.70% (244) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3420c | - | 9e-32 | 40.20% (204) | short chain dehydrogenase |
| M. marinum M | MMAR_4755 | - | 4e-33 | 38.24% (238) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_0895 | - | 1e-34 | 36.15% (260) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5235 | - | 2e-38 | 45.41% (196) | short chain dehydrogenase family protein |
| M. thermoresistible (build 8) | TH_2445 | - | 5e-38 | 46.94% (196) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_3400 | - | 9e-33 | 36.51% (241) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_01740|M.leprae_Br4923 -----MVKTAQYFVGKRCFVTGAASGIGRATALRLAAYGAELYLTDRHCD
MUL_3400|M.ulcerans_Agy99 ------MAQRRYFAGKRCLVTGAASGIGRATALRLAEQGAELYLTDRDGD
MAB_3420c|M.abscessus_ATCC_199 --------MAYEFTGKRCFVTGAASGIGRATALRLGREGAELFLTDRAAD
MSMEG_5235|M.smegmatis_MC2_155 ----------MSFTGKTAIVTGAGSGIGATLCRALVDAGADVLCTDVDAD
TH_2445|M.thermoresistible__bu ----------MTFAGKHALVTGAGSGIGAALCRALAAAGAEVMCTDIDG-
Mflv_3370|M.gilvum_PYR-GCK ----MASTDTAQLSGKVAFVTGGASGIGAALVAKLATAGTEVWIADRQIA
Mb0874c|M.bovis_AF2122/97 -----MDGFP----GRGAVITGGASGIGLATGTEFARRGARVVLGDVDKP
Rv0851c|M.tuberculosis_H37Rv -----MDGFP----GRGAVITGGASGIGLATGTEFARRGARVVLGDVDKP
MAV_0895|M.avium_104 -----MDGFLSGFDGRAAVVTGGASGIGLATATEFARRGARLVLSDVDQP
MMAR_4755|M.marinum_M -----MDGFA----GRGAVITGGASGIGLATATEFARRGAKVVLADIDKP
Mvan_0852|M.vanbaalenii_PYR-1 MVSNHFR-------GKVCVVTGAASGIGLAVGAALLDHGATVVLADFDAD
*: ..:**..**** : : *: : *
MLBr_01740|M.leprae_Br4923 GLAQTVSEARALGAQVPEYRALDISDYDEVAAFGADIHARHPSMDIVMNI
MUL_3400|M.ulcerans_Agy99 GLAQTVSAARALGAQVPEHRVLDISDYDEVAAFAADIHASHPSMDVVLNI
MAB_3420c|M.abscessus_ATCC_199 GLESVVEEIKAAGGKVSAYRALDISDHEAVAAFASDILATHPSMDVLLNI
MSMEG_5235|M.smegmatis_MC2_155 AAARTAAARTAA-ALGARSARLDVTDAAAVQTTVDDVVARAGRLDLMFNN
TH_2445|M.thermoresistible__bu ----AAAERTAQ-SIGGRWAALDVTDAAAVQAAVDGVVDRAGRLDLLFNN
Mflv_3370|M.gilvum_PYR-GCK PAEELAAQLRAT-GAVAHAVELDVRDPAAFAHAVHTAVDTSGRLDYLFNN
Mb0874c|M.bovis_AF2122/97 GLRQAVNHLRAE-GFDVHGVMCDVRHREEVTHLADEAFRLLGHFDVVFSN
Rv0851c|M.tuberculosis_H37Rv GLRQAVNHLRAE-GFDVHSVMCDVRHREEVTHLADEAFRLLGHVDVVFSN
MAV_0895|M.avium_104 ALEQAVNGLRGQ-GFDAHGVVCDVRHLDEMVRLADEAFRLLGGVDVVFSN
MMAR_4755|M.marinum_M GLERSVEHLRGK-GFDAHGVMCDVRHLSEVNHLAAESARLLGQIDVVFSN
Mvan_0852|M.vanbaalenii_PYR-1 RLTTVAEGLCMH-DGRVHSAVVDVTVQEAVQALVEGAARAHGSLDFLFNN
. *: . .* ::.
MLBr_01740|M.leprae_Br4923 AGVSVWGTVDRLTHDQWSNVVAINLMGPIHIIETFVPAILAAGRGGHLVN
MUL_3400|M.ulcerans_Agy99 AGISAWGTVDRLSHEHWSKMVAVNLMGPIHVIETFVPPMVAAGRGGHLVN
MAB_3420c|M.abscessus_ATCC_199 AGISVWGSVDRLSHQQWRSVVDINLMGPIHVIESFVPAMIKARKGGQIVN
MSMEG_5235|M.smegmatis_MC2_155 AGIVWGGDTELLTLDQWNAIIDVNIRGVVHGVAAAYPQMIRQGHG-HIVN
TH_2445|M.thermoresistible__bu AGIVWGGDTELLTLDQWNAIIDVNIRGVVHGVAAAYPLMVRQGHG-HIVN
Mflv_3370|M.gilvum_PYR-GCK AGIGIGGEVDTLTLDDWTDIIDVNLKGVIHGVHAAYPQMIRQGSG-HIVN
Mb0874c|M.bovis_AF2122/97 AGIVVGGPIVEMTHDDWRWVIDVDLWGSIHTVEAFLPRLLEQGTGGHVVF
Rv0851c|M.tuberculosis_H37Rv AGIVVGGPIVEMTHDDWRWVIDVDLWGSIHTVEAFLPRLLEQGTGGHVVF
MAV_0895|M.avium_104 AGIVVAGPLAQMNHDDWRWVIDIDLWGSIHAVEAFLPRLLEQGTGGHIAF
MMAR_4755|M.marinum_M AGIVVAGPIADMTHDDWRWVIDIDLWGSIHTVEAFLPKLLEHG--GHIAF
Mvan_0852|M.vanbaalenii_PYR-1 AGVGGTMPIGVATLEQWRRIVDLNLWGVIYGVHAAVPIMVRQGS-GHIVN
**: . :.* :: ::: * :: : : * :: ::.
MLBr_01740|M.leprae_Br4923 VSSAAGLVALPWHAAYSASKYGLRGLSEVLRFDLARYRIGVSVVVPGAVR
MUL_3400|M.ulcerans_Agy99 VSSAAGLVALPWHAAYSASKYGLRGLSEVLRFDLARHRIGVSVVVPGAVD
MAB_3420c|M.abscessus_ATCC_199 VSSAAGIVALPWHAAYSASKYGLRGVSEVLRFDLARYKIGVTVVVPGAVN
MSMEG_5235|M.smegmatis_MC2_155 TASMAGLAAAGQLTSYVMSKHAVVGLSLALRSEAAAHGVGVLAVCPAAVE
TH_2445|M.thermoresistible__bu TASMAGLAAAGQLTSYVMTKHAVVGLSLALRSEAAAHGVGVLAVCPAAVE
Mflv_3370|M.gilvum_PYR-GCK TASMAGLITTSAQGGYSATKHAVVALSKTMRVEAATHGVRVSVLCPGVVR
Mb0874c|M.bovis_AF2122/97 TASFAGLVPNAGLGAYGVAKYGVVGLAETLAREVTADGIGVSVLCPMVVE
Rv0851c|M.tuberculosis_H37Rv TASFAGLVPNAGLGAYGVAKYGVVGLAETLAREVTADGIGVSVLCPMVVE
MAV_0895|M.avium_104 TASFAGLVPNAGLGTYGVAKYGVVGLAETLAREVKPNGIGVSVLCPMVVE
MMAR_4755|M.marinum_M TASFAGLVPNAGLGAYGVAKYGVVGLAETLSREMKDRGVGVTVLCPMVVE
Mvan_0852|M.vanbaalenii_PYR-1 TASLAGLVPFPYQSLYCTTKYGVVGLSESLRYELAADGVRVSVVCPGNVV
.:* **: . * :*:.: .:: : : : * .: * *
MLBr_01740|M.leprae_Br4923 TPLVDTIEIAGVDRQDPRFSRW-VDRFRGHAVSAERAAEKILAGVAKNRY
MUL_3400|M.ulcerans_Agy99 TPLVDTVEIAGVDREDPQVNRW-VNRFSGHAVSPEKAAEKILAGVARNRY
MAB_3420c|M.abscessus_ATCC_199 TPLVRTVDIAGVDRENENVARW-IGRFVGHAVSPEKVADRILYGIRHNRY
MSMEG_5235|M.smegmatis_MC2_155 TPILDKGAVGG--FVGRDYFLR-GQGMK-TAYDPDRLAADTLRAIERNKA
TH_2445|M.thermoresistible__bu TPLLDKGAVGG--FLGRDYFIR-GQGVR-SAYDPDRLASDTLRAVQRNKA
Mflv_3370|M.gilvum_PYR-GCK TPILTGGAFGRNKSVSREDQLK-LGEAL-RPMDAGVFAQKALRGVLRNEA
Mb0874c|M.bovis_AF2122/97 TNLVANSERIRGAACAQSST---TGSPGPLPLQDDNLGVDDIAQLTADAI
Rv0851c|M.tuberculosis_H37Rv TNLVANSERIRGAACAQSST---TGSPGPLPLQDDNLGVDDIAQLTADAI
MAV_0895|M.avium_104 TKLVSNSERIRGADYGMSATP--EGAFGPLPTQDESVSADDVARLTADAI
MMAR_4755|M.marinum_M TRLVSNSERIRGADYGLSSAPQVTGDMGSLPAQDDSLSVDELARLTADAI
Mvan_0852|M.vanbaalenii_PYR-1 SRIFGTPILGRPVDAAPPEDS-IPAEEAARLILAGVAAGEGIIAFPEKPR
: :. . : . .
MLBr_01740|M.leprae_Br4923 LIYTSHDIRVLYGFKRLAWWPYGVVMKQVNVVFTRALRPSPATMRCVEPI
MUL_3400|M.ulcerans_Agy99 LVYTSADIRALYAFKRLAWWPYSVAMRQVNVIFTRALKPKPVALPQRE--
MAB_3420c|M.abscessus_ATCC_199 LVYTSVDIRALYLFKRTLWWPYSVAMRVANYAFTNALRPGFQKTGSAS--
MSMEG_5235|M.smegmatis_MC2_155 LLVKPRRAHASWLLARLAPG----LMQRLSVRFVAAQRASQARTANH---
TH_2445|M.thermoresistible__bu LLVKPRVAHAGWLFARLAPG----LMQRASIRFVERQRAGQRAAQVGR--
Mflv_3370|M.gilvum_PYR-GCK IIVVPRWWKALWYLERLSPA----LSMRTAAVALKRTREVQSAVPLQ---
Mb0874c|M.bovis_AF2122/97 LANRLYVLPHAASRASIRRR-----FERIDRTFDEQAAEGWRH-------
Rv0851c|M.tuberculosis_H37Rv LANRLYVLPHAASRASIRRR-----FERIDRTFDEQAAEGWRH-------
MAV_0895|M.avium_104 LANRLYILPHAAARESIRRR-----FERIDRTFDEQAAEGWTH-------
MMAR_4755|M.marinum_M LANRLYVLPHEGARTSIQRR-----FARIDRTFDEQAAEGWRH-------
Mvan_0852|M.vanbaalenii_PYR-1 ELWRSYAESPDSVEGYLKEV-----ARQRRAAFATGDAQALFRATQSAGG
MLBr_01740|M.leprae_Br4923 KVGVYSQPRSGEGGKSGN
MUL_3400|M.ulcerans_Agy99 ------------------
MAB_3420c|M.abscessus_ATCC_199 ------SPRDS-------
MSMEG_5235|M.smegmatis_MC2_155 ------------------
TH_2445|M.thermoresistible__bu ------------------
Mflv_3370|M.gilvum_PYR-GCK ------------------
Mb0874c|M.bovis_AF2122/97 ------------------
Rv0851c|M.tuberculosis_H37Rv ------------------
MAV_0895|M.avium_104 ------------------
MMAR_4755|M.marinum_M ------------------
Mvan_0852|M.vanbaalenii_PYR-1 T-----------------