For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSFTGKTAIVTGAGSGIGATLCRALVDAGADVLCTDVDADAAARTAAARTAAALGARSARLDVTDAAAVQ TTVDDVVARAGRLDLMFNNAGIVWGGDTELLTLDQWNAIIDVNIRGVVHGVAAAYPQMIRQGHGHIVNTA SMAGLAAAGQLTSYVMSKHAVVGLSLALRSEAAAHGVGVLAVCPAAVETPILDKGAVGGFVGRDYFLRGQ GMKTAYDPDRLAADTLRAIERNKALLVKPRRAHASWLLARLAPGLMQRLSVRFVAAQRASQARTANH
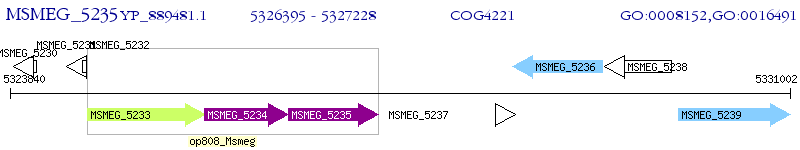
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5235 | - | - | 100% (277) | short chain dehydrogenase family protein |
| M. smegmatis MC2 155 | MSMEG_4280 | - | 5e-35 | 36.23% (265) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2291 | - | 8e-33 | 34.05% (279) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3112 | - | 3e-35 | 37.55% (269) | short-chain type dehydrogenase/reductase |
| M. gilvum PYR-GCK | Mflv_2070 | - | 1e-107 | 74.26% (272) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv3085 | - | 3e-35 | 37.55% (269) | short-chain type dehydrogenase/reductase |
| M. leprae Br4923 | MLBr_01094 | - | 8e-33 | 33.58% (274) | short chain alcohol dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4022c | - | 6e-36 | 36.33% (267) | oxidoreductase |
| M. marinum M | MMAR_2600 | - | 3e-41 | 36.63% (273) | short-chain dehydrogenase |
| M. avium 104 | MAV_1384 | - | 5e-33 | 35.38% (260) | short chain alcohol dehydrogenase |
| M. thermoresistible (build 8) | TH_2445 | - | 1e-125 | 81.92% (271) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_3160 | - | 4e-41 | 36.63% (273) | short-chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_4644 | - | 1e-112 | 76.21% (269) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5235|M.smegmatis_MC2_155 ----------MSFTGKTAIVTGAGSGIGATLCRALVDAGADVLCTDVDAD
TH_2445|M.thermoresistible__bu ----------MTFAGKHALVTGAGSGIGAALCRALAAAGAEVMCTDIDG-
Mflv_2070|M.gilvum_PYR-GCK ------MSSTADFAGKQALLTGGGSGIGAALCRALDRAGAHVVCTDIDE-
Mvan_4644|M.vanbaalenii_PYR-1 ----------MRFTGKHAIITGGGSGIGAALCRALDRAGAQVVCTDIDE-
MAB_4022c|M.abscessus_ATCC_199 ------MVKLNQLQPQLVVVTGAGSGIGRATAIRFAKRGAHVVVSDMDLD
MMAR_2600|M.marinum_M MFKLNSGTNTSDLAGRVVAITGAGSGIGRELALLCAQRGADLALCDINDT
MUL_3160|M.ulcerans_Agy99 MFKLNSGTNTSDLAGRVVAITGAGSGIGRELALLCAQRGADLALCDINDT
Mb3112|M.bovis_AF2122/97 ---------MSSFEGKVAVITGAGSGIGRALALNLSEKRAKLALSDVDTD
Rv3085|M.tuberculosis_H37Rv ---------MSSFEGKVAVITGAGSGIGRALALNLSEKRAKLALSDVDTD
MLBr_01094|M.leprae_Br4923 ---------MEGFAGKVAVVTGAGSGIGQALAIELARSGAKLAISDVDGE
MAV_1384|M.avium_104 ---------MQGFAGKVAVVTGAGSGIGQALAVELARSGAKVAISDVDLE
: : . :**.***** . *.: *::
MSMEG_5235|M.smegmatis_MC2_155 AAARTAAARTAAALG--ARSARLDVTDAAAVQTTVDDVVARAGRLDLMFN
TH_2445|M.thermoresistible__bu ----AAAERTAQSIG--GRWAALDVTDAAAVQAAVDGVVDRAGRLDLLFN
Mflv_2070|M.gilvum_PYR-GCK ----LAAERVVSTLSSRARAARLDVTDSAAVQAAVDDVEARAGRLDLMFN
Mvan_4644|M.vanbaalenii_PYR-1 ----AAAGAVVSTLSPRARAARLDVTDAAAVQGVVDDVVARTGRLDLIFN
MAB_4022c|M.abscessus_ATCC_199 S--AHATTAMIQADGNTASAVQLDVTDPDQWETFARDVLADHGVADVLVN
MMAR_2600|M.marinum_M A--VADTAQTARGFGHDVITRRVDVSDPEQMTAFADATLGHFGGVDLLVN
MUL_3160|M.ulcerans_Agy99 A--VADTAQTARGFGHDVITRRVDVSDPEQMTAFADATLGHFGGVDLLVN
Mb3112|M.bovis_AF2122/97 G--LAKTVRLAQALGAQVKSDRLDVAEREAVLAHADAVVAHFGTVHQVYN
Rv3085|M.tuberculosis_H37Rv G--LAKTVRLAQALGAQVKSDRLDVAEREAVLAHADAVVAHFGTVHQVYN
MLBr_01094|M.leprae_Br4923 G--LAQTEGQLKAIGASARTDRLDVTEREAFLTYADVVHENFGKVNQIYN
MAV_1384|M.avium_104 G--LAHTEEQLKAIGAQYKADRLDVTEREAFLAYADAVKEHFGKVNQIYN
: . :**:: . . * . : *
MSMEG_5235|M.smegmatis_MC2_155 NAGIVWGGDTELLTLDQWNAIIDVNIRGVVHGVAAAYPQMIRQGHG-HIV
TH_2445|M.thermoresistible__bu NAGIVWGGDTELLTLDQWNAIIDVNIRGVVHGVAAAYPLMVRQGHG-HIV
Mflv_2070|M.gilvum_PYR-GCK NAGIVFGGETELLTLDQWNAIIDVNIRGVVHGVAAAYPLMLRQGHG-HIV
Mvan_4644|M.vanbaalenii_PYR-1 NAGIVWGGDTELLTLDQWNAIIDINLRGVVHGVAAAYPQMLRQGHG-HIV
MAB_4022c|M.abscessus_ATCC_199 NAGIALGGPFLKLTPADWDRLLSVNLMGVVHGCRVFGEQMVERGSG-HIV
MMAR_2600|M.marinum_M NAGVGLIGGFLDTSRKDWDWLVSINVMGVVHGCEAFLPAMIESGRGGHVV
MUL_3160|M.ulcerans_Agy99 NAGVGLIGGFLDTSRKDWDWLVSINVMGVVHGCEAFLPAMIESGRGGHVV
Mb3112|M.bovis_AF2122/97 NAGIAYNGNVDKSEFKDIERIIDVDFWGVVNGTKAFLPHVIASGDG-HIV
Rv3085|M.tuberculosis_H37Rv NAGIAYNGNVDKSEFKDIERIIDVDFWGVVNGTKAFLPHVIASGDG-HIV
MLBr_01094|M.leprae_Br4923 NAGIAFTGDVEVSHFKDIERVMDVDYWGVVNGTKAFLPYLISSGDG-HVI
MAV_1384|M.avium_104 NAGIAFTGDVEVSQFKDIERVMDVDFWGVVNGTKAFLPHLIASGDG-HVV
***: * : : ::.:: ***:* . :: * * *::
MSMEG_5235|M.smegmatis_MC2_155 NTASMAGLAAAGQLTSYVMSKHAVVGLSLALRSEAAAHG--VGVLAVCPA
TH_2445|M.thermoresistible__bu NTASMAGLAAAGQLTSYVMTKHAVVGLSLALRSEAAAHG--VGVLAVCPA
Mflv_2070|M.gilvum_PYR-GCK NTASMAGLAAAGHLTSYVATKHAVVGLSLALRSEALSRG--IGVLAVCPA
Mvan_4644|M.vanbaalenii_PYR-1 NTASMAGLTAAGQVTSYVATKHAVVGLSMALRSEAVPRG--VGVLVVCPA
MAB_4022c|M.abscessus_ATCC_199 NIASAAAFTPTAVMAPYSVSKAGVKMLTECLRLELGPKG--VGVSAICPG
MMAR_2600|M.marinum_M NLSSAAGLLANSALSAYSATKFAVLGLSEALRIELEPHR--IGVTAICPG
MUL_3160|M.ulcerans_Agy99 NLSSAAGLLANPALSAYSATKFAVLGLSEALRIELEPHR--IGVTAICPG
Mb3112|M.bovis_AF2122/97 NISSLFGLIAVPGQSAYNAAKFAVRGFTEALRQEMLVARHPVKVTCVHPG
Rv3085|M.tuberculosis_H37Rv NISSLFGLIAVPGQSAYNAAKFAVRGFTEALRQEMLVARHPVKVTCVHPG
MLBr_01094|M.leprae_Br4923 NISSVFGLFSVPGQAAYNSAKFAVRGFTEALREEMALAGRPVNVTTVYPG
MAV_1384|M.avium_104 NVSSVFGLFSVPGQAAYNSAKFAVRGFTEALRQEMAAAGHPVAVTTVHPG
* :* .: . :.* :* .* :: .** * : * : *.
MSMEG_5235|M.smegmatis_MC2_155 AVETPILDKGAVGGFV------GRDYFLRGQGMKT-----AYDPDRLAAD
TH_2445|M.thermoresistible__bu AVETPLLDKGAVGGFL------GRDYFIRGQGVRS-----AYDPDRLASD
Mflv_2070|M.gilvum_PYR-GCK AVETPILDKGAVGSFG------GRDFFLRAQGGK------AYDADRLARE
Mvan_4644|M.vanbaalenii_PYR-1 AVETPILDKGAVGGFV------GRDYFLQTQGGR------AYDADRLARD
MAB_4022c|M.abscessus_ATCC_199 FINTNIGLNGITVGVDQEMVQRGKEIMRRAQEFAAHLPFSPMSPDLVARA
MMAR_2600|M.marinum_M VINTAITKASPIRGAG---DTEGRRQRLTVTYQRR-----GYTPARAARN
MUL_3160|M.ulcerans_Agy99 VINTAITKASPIRGAG---DTEGRRQRLTVTYQRR-----GYTPARAARN
Mb3112|M.bovis_AF2122/97 GIKTAVARNATVADGE------DQQTFAEFFDRRLA----LHSPEMAAKT
Rv3085|M.tuberculosis_H37Rv GIKTAVARNATVADGE------DQQTFAEFFDRRLA----LHSPEMAAKT
MLBr_01094|M.leprae_Br4923 GIKTAIARNATAAEGL------DVSKIASRFDTWVA----HTSPQHAARI
MAV_1384|M.avium_104 GIKTAIARNATAAEGL------DQAELAKLFDKRLA----KTTPQRAAQI
::* : . . . *
MSMEG_5235|M.smegmatis_MC2_155 TLRAIERNKALLVKPRRAHASWLLARLAPGLMQRLSVRFVAAQRASQAR-
TH_2445|M.thermoresistible__bu TLRAVQRNKALLVKPRVAHAGWLFARLAPGLMQRASIRFVERQRAGQRA-
Mflv_2070|M.gilvum_PYR-GCK VLDAITRNKAVLVKPRVAHAQWLLNRLAPTGLNRLASRFLAAQQSSRVN-
Mvan_4644|M.vanbaalenii_PYR-1 TLRAVERNKAILVKPTVAQAQWWFARLAPGLMNRMAMRFVTGQRTRQVDR
MAB_4022c|M.abscessus_ATCC_199 VTRAVRYDLAVVPVRTEAWLGYVLGRVAPGINRRTTQAFSIERAERWGAW
MMAR_2600|M.marinum_M ILRAVERNKAVAPVAAEAHVMYVLSRIMPPLARWVSAQMAKAAK------
MUL_3160|M.ulcerans_Agy99 ILRAVERNKAVAPVAAEAHVMYVLSRIMPPLARWVSAQMAKAAK------
Mb3112|M.bovis_AF2122/97 IVNGVAKGQARVVVGLEAKAVDVLARIMG-SSYQRLVAAGVAKFFPWAK-
Rv3085|M.tuberculosis_H37Rv IVNGVAKGQARVVVGLEAKAVDVLARIMG-SSYQRLVAAGVAKFFPWAK-
MLBr_01094|M.leprae_Br4923 ILKAVRKKKARVLVGPDAKVANVVVRFSGGAGYQRLFAQVASRLILNQR-
MAV_1384|M.avium_104 ILDAVRKKKARVLVGSDAKALDILVRLTG-SGYQRLFGPVMSRLLPN---
.: * * . *.
MSMEG_5235|M.smegmatis_MC2_155 -----TANH-------------------
TH_2445|M.thermoresistible__bu -----AQVGR------------------
Mflv_2070|M.gilvum_PYR-GCK -------SG-------------------
Mvan_4644|M.vanbaalenii_PYR-1 YPSSAADTG-------------------
MAB_4022c|M.abscessus_ATCC_199 ALGKLDDLNLLSRQGGVEEDRKPTAARR
MMAR_2600|M.marinum_M ----------------------------
MUL_3160|M.ulcerans_Agy99 ----------------------------
Mb3112|M.bovis_AF2122/97 ----------------------------
Rv3085|M.tuberculosis_H37Rv ----------------------------
MLBr_01094|M.leprae_Br4923 ----------------------------
MAV_1384|M.avium_104 ----------------------------