For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTTNADRKRPGRPPGTSDTRDRILECARELFARNGITNTSIRSIAAGAGVDPALVHHYFGTKTQLFAAAI EIPIDPMSIIGPLKEVPVDQLGRALPSLLLPLWDSELGKGFIATLRSILAGSDVSMVRSFLQDLIATEVG GRVDDPPGSGRVRVQFVASQLVGIVMARYILELEPFKSLPVEQIVDTVAPNLQRYLTGELPQFSGD
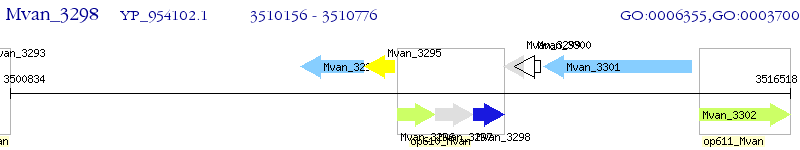
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3298 | - | - | 100% (206) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_2569 | - | 7e-26 | 37.24% (196) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_0641 | - | 3e-25 | 34.69% (196) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1711c | - | 4e-82 | 72.31% (195) | hypothetical protein Mb1711c |
| M. gilvum PYR-GCK | Mflv_0253 | - | 5e-24 | 34.54% (194) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv1685c | - | 4e-82 | 72.31% (195) | hypothetical protein Rv1685c |
| M. leprae Br4923 | MLBr_00949 | - | 8e-05 | 33.80% (71) | hypothetical protein MLBr_00949 |
| M. abscessus ATCC 19977 | MAB_2351c | - | 3e-61 | 54.31% (197) | TetR family transcriptional regulator |
| M. marinum M | MMAR_2484 | - | 2e-76 | 63.68% (201) | hypothetical protein MMAR_2484 |
| M. avium 104 | MAV_3088 | - | 4e-83 | 73.50% (200) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_3765 | - | 8e-92 | 80.60% (201) | transcriptional regulator |
| M. thermoresistible (build 8) | TH_3542 | - | 5e-22 | 32.96% (179) | PUTATIVE transcriptional regulator, TetR family |
| M. ulcerans Agy99 | MUL_1664 | - | 2e-71 | 61.54% (195) | hypothetical protein MUL_1664 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0253|M.gilvum_PYR-GCK MPPPAQT---GRRRGRRKGDPVSRETVLAAAKGRFARDGFEKTTLRAIAE
TH_3542|M.thermoresistible__bu --------------------------VLAAAKRRFAQDGYEKTTLRAIAR
Mb1711c|M.bovis_AF2122/97 MAAPDNS---RRRPGRPAGSSDTRERILSSARELFAHNGIDRTSIRAVAA
Rv1685c|M.tuberculosis_H37Rv MAAPDNS---RRRPGRPAGSSDTRERILSSARELFAHNGIDRTSIRAVAA
MMAR_2484|M.marinum_M METTNTG---RRRPGRPTGTSNTREHILTCARELFALNGLDRTSVRSVAA
MUL_1664|M.ulcerans_Agy99 METTNTG---RRRPGRPTGTSNTREHILTCARELFALNGLDRTSVRSVAA
Mvan_3298|M.vanbaalenii_PYR-1 MTTNAD----RKRPGRPPGTSDTRDRILECARELFARNGITNTSIRSIAA
MSMEG_3765|M.smegmatis_MC2_155 MTATSDTGRARKRPGRPPGASDTRDRILASARELFARNGFDKTSIRAVAA
MAV_3088|M.avium_104 MTTTGTT---RRRRGRPAGSSGSRERILASARELFARNGIRNTSIRAVAA
MAB_2351c|M.abscessus_ATCC_199 MS--------SKRPGRRPGTSSARDDILASARKLFSLNGIDKTSIRAIAS
MLBr_00949|M.leprae_Br4923 MTTS-------ARTHRWAKTGAIQRRILDAAIEVFATRSFTTATMTDAMT
:* .* *: . :::
Mflv_0253|M.gilvum_PYR-GCK DAGVDASMVLYLFGSKEALFRESLRLILDPGVLIEALSGG-EADIGTRMV
TH_3542|M.thermoresistible__bu DAHVDASMVLYLFGSKAELFRESLELILDPEMLVAAMTGD-DGDVGTRMV
Mb1711c|M.bovis_AF2122/97 KAGVDAALVHHYFGTKQQLFAAAIHIPIDPMVIIGPIREAPVEELGYKLP
Rv1685c|M.tuberculosis_H37Rv KAGVDAALVHHYFGTKQQLFAAAIHIPIDPMVIIGPIREAPVEELGYKLP
MMAR_2484|M.marinum_M AAGVDASLVHHYYGTKQQLFAAAIQIPIDPTVVLEQIVETPVDELGLKLP
MUL_1664|M.ulcerans_Agy99 AAGVDASLVHHYYGTKQQLFAAAIQIPIDPTVVLEQIVETPVDELGLKIP
Mvan_3298|M.vanbaalenii_PYR-1 GAGVDPALVHHYFGTKTQLFAAAIEIPIDPMSIIGPLKEVPVDQLGRALP
MSMEG_3765|M.smegmatis_MC2_155 GADVDPALVHHYFGTKTQLFAAAIHIPIDPLEVIGPLREVPVEQIGHTLP
MAV_3088|M.avium_104 AAGVDSALVHHYFGTKEKLFAAAVQIPIDPMQVIGPLREVPVDDLGYALP
MAB_2351c|M.abscessus_ATCC_199 DAGVDPALVHHYFGTKLDLFREVVQLPVDPSVVLQPLRDVPVDELGVTIP
MLBr_00949|M.leprae_Br4923 GSGAIIGSIYHHFGGKRKLFLAIFEQMGR--------QRRPPHRRGDATS
: . . : : :* * ** .. *
Mflv_0253|M.gilvum_PYR-GCK RAYLGVWESPESAESMRTMLASATSNPDAREAFRGFIQDYVLAAVVGTLG
TH_3542|M.thermoresistible__bu RVYLQIWESPDTGPTIRAMLQSATSNTDAREAFRSFLREYLLTAVSGVLG
Mb1711c|M.bovis_AF2122/97 SLLLPIWDS-ELGAGLIATLRSLISGSDV-GLARSFLEEVVTVELGSRVD
Rv1685c|M.tuberculosis_H37Rv SLLLPIWDS-ELGAGLIATLRSLISGSDV-GLARSFLEEVVTVELGSRVD
MMAR_2484|M.marinum_M SVLLPLWDS-ELGTGLVSTLRALMAGTES-NLARSFLQEVVTSEVGSRVD
MUL_1664|M.ulcerans_Agy99 SVPLPLWDS-ELSTGLVATLRALMAGTES-NLARSFLQEVVTSEVGSRVD
Mvan_3298|M.vanbaalenii_PYR-1 SLLLPLWDS-ELGKGFIATLRSILAGSDV-SMVRSFLQDLIATEVGGRVD
MSMEG_3765|M.smegmatis_MC2_155 RLLLPLWDS-ELGKGFIATLRSILAGNDV-SLVRSFLQEVIVGEVGSRVD
MAV_3088|M.avium_104 SMLLPLWDS-EVGAAFIATLRSILAGEEI-SLFRTFIQDVIGVEVGPRVD
MAB_2351c|M.abscessus_ATCC_199 RLIIALWDS-ELGANMLAVFRSALTGADD-GLVRVFFREVLVNIIAERVD
MLBr_00949|M.leprae_Br4923 GPIRPSGDR------SAAGIRSPRTGPTS--------------RRCGKTG
: : : : :. .
Mflv_0253|M.gilvum_PYR-GCK G-DDRARLRAMLAASSLVGTAMMRYLMEIPPLSELATEDVIRLVAPTVTR
TH_3542|M.thermoresistible__bu G-GEQARLRAELAATGLVGTAMLRYIMRVPPLAELSTEEVVRMLAPTVNR
Mb1711c|M.bovis_AF2122/97 NPPGTGKIRTQFVASQLMGVVMARYIVRIEPFASLPAEQIVQTIAPNLQR
Rv1685c|M.tuberculosis_H37Rv NPPGTGKIRTQFVASQLMGVVMARYIVRIEPFASLPAEQIVQTIAPNLQR
MMAR_2484|M.marinum_M SPTGTGRIRAQFVASQLLGVVMARHIVKIEPFASLPADQVAQAIAPTLQR
MUL_1664|M.ulcerans_Agy99 SPTGTGRIRAQFVTSQLLGVVMARHIVKIEPFASLPADQVAQAIAPTLQR
Mvan_3298|M.vanbaalenii_PYR-1 DPPGSGRVRVQFVASQLVGIVMARYILELEPFKSLPVEQIVDTVAPNLQR
MSMEG_3765|M.smegmatis_MC2_155 NPAGSGRIRVQYVASQLVGVVMARYILELEPFKSLPVEQIAETIGPNLQR
MAV_3088|M.avium_104 NPAGSGVIRIQFVASQLVGVVMARYILELEPFASLPPEQIAATIAPNLQR
MAB_2351c|M.abscessus_ATCC_199 SPPGSGVLRAEFAITQMAGILVGRYIMAIEPLASLTAEQIALTVGPNIQR
MLBr_00949|M.leprae_Br4923 TRPGCCRPETHLQASR-------------SPVATA---------------
. : *.
Mflv_0253|M.gilvum_PYR-GCK YLTAPADELGLPG------
TH_3542|M.thermoresistible__bu YLTADADELGLPSSLTAGS
Mb1711c|M.bovis_AF2122/97 YLTGELPDDLAP-------
Rv1685c|M.tuberculosis_H37Rv YLTGELPDDLAP-------
MMAR_2484|M.marinum_M YLTGELPEEIAS-------
MUL_1664|M.ulcerans_Agy99 YLNR---------------
Mvan_3298|M.vanbaalenii_PYR-1 YLTGELPQFSGD-------
MSMEG_3765|M.smegmatis_MC2_155 YLTGELPGFGALD------
MAV_3088|M.avium_104 YLTGDLPDGLAP-------
MAB_2351c|M.abscessus_ATCC_199 YLTGALPSITP--------
MLBr_00949|M.leprae_Br4923 -------------------