For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PPPAQTGRRRGRRKGDPVSRETVLAAAKGRFARDGFEKTTLRAIAEDAGVDASMVLYLFGSKEALFRESL RLILDPGVLIEALSGGEADIGTRMVRAYLGVWESPESAESMRTMLASATSNPDAREAFRGFIQDYVLAAV VGTLGGDDRARLRAMLAASSLVGTAMMRYLMEIPPLSELATEDVIRLVAPTVTRYLTAPADELGLPG*
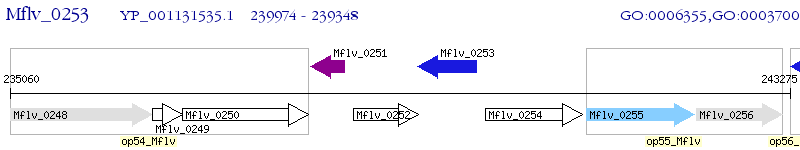
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0253 | - | - | 100% (208) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_0297 | - | 3e-12 | 29.74% (195) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_4147 | - | 1e-07 | 45.07% (71) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1711c | - | 4e-26 | 32.50% (200) | hypothetical protein Mb1711c |
| M. tuberculosis H37Rv | Rv1685c | - | 4e-26 | 32.50% (200) | hypothetical protein Rv1685c |
| M. leprae Br4923 | MLBr_02568 | - | 5e-05 | 28.10% (153) | putative TetR-family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_2351c | - | 4e-26 | 34.36% (195) | TetR family transcriptional regulator |
| M. marinum M | MMAR_3397 | - | 3e-27 | 33.16% (187) | putative regulatory protein |
| M. avium 104 | MAV_3088 | - | 2e-26 | 36.87% (198) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_2182 | - | 8e-29 | 37.56% (197) | transcriptional regulator |
| M. thermoresistible (build 8) | TH_3542 | - | 9e-71 | 69.57% (184) | PUTATIVE transcriptional regulator, TetR family |
| M. ulcerans Agy99 | MUL_2781 | - | 2e-25 | 33.67% (199) | hypothetical protein MUL_2781 |
| M. vanbaalenii PYR-1 | Mvan_0641 | - | 2e-85 | 75.71% (210) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1711c|M.bovis_AF2122/97 ----------------------------------------MAAPDNSRRR
Rv1685c|M.tuberculosis_H37Rv ----------------------------------------MAAPDNSRRR
MUL_2781|M.ulcerans_Agy99 MQPTTTHPAQARRLRSSGKLLGARDVSAHNKPRNVDNTRGGNAPANERRR
MAV_3088|M.avium_104 ----------------------------------------MTTTGTTRRR
MAB_2351c|M.abscessus_ATCC_199 ---------------------------------------------MSSKR
MMAR_3397|M.marinum_M --------------------------------------------------
TH_3542|M.thermoresistible__bu --------------------------------------------------
Mvan_0641|M.vanbaalenii_PYR-1 -----------------------------------------MPRPGETAR
Mflv_0253|M.gilvum_PYR-GCK -----------------------------------------MPPPAQTGR
MSMEG_2182|M.smegmatis_MC2_155 ---------------------------------------------MAENR
MLBr_02568|M.leprae_Br4923 -------------------------------------------------M
Mb1711c|M.bovis_AF2122/97 -PGRPAG-SSDTRERILSSARELFAHNGIDRTSIRAVAAKAGVDAALVHH
Rv1685c|M.tuberculosis_H37Rv -PGRPAG-SSDTRERILSSARELFAHNGIDRTSIRAVAAKAGVDAALVHH
MUL_2781|M.ulcerans_Agy99 RPGRLRGNSSDTRQRILASARELFASNGIDRTSIRAVAAAADVDAALVHH
MAV_3088|M.avium_104 -RGRPAG-SSGSRERILASARELFARNGIRNTSIRAVAAAAGVDSALVHH
MAB_2351c|M.abscessus_ATCC_199 -PGRRPG-TSSARDDILASARKLFSLNGIDKTSIRAIASDAGVDPALVHH
MMAR_3397|M.marinum_M -MGRRRG-NPDTRNAIVTTARRLFAESGYEQTSVRDIASAAGVDPALIRH
TH_3542|M.thermoresistible__bu ---------------VLAAAKRRFAQDGYEKTTLRAIARDAHVDASMVLY
Mvan_0641|M.vanbaalenii_PYR-1 RRGRRQG-EPVSKEAVLAAAKTRFARDGYEKTTLRAIADDARVDASMVLY
Mflv_0253|M.gilvum_PYR-GCK RRGRRKG-DPVSRETVLAAAKGRFARDGFEKTTLRAIAEDAGVDASMVLY
MSMEG_2182|M.smegmatis_MC2_155 RSGRWRT-GQQNKQRILDAARDHFMRSGYEKATVRGIAAEAGVDVAMVYY
MLBr_02568|M.leprae_Br4923 AGGAKRIPRAVREQQMLDAAVQMFSVNGYHKTSMDAIAAEAQISKPMLYL
:: :* * .* .::: :* * :. .::
Mb1711c|M.bovis_AF2122/97 YFGTKQQLFAAAIHIPIDPMVIIGPIREA----PVEELGYKLPSLLLPIW
Rv1685c|M.tuberculosis_H37Rv YFGTKQQLFAAAIHIPIDPMVIIGPIREA----PVEELGYKLPSLLLPIW
MUL_2781|M.ulcerans_Agy99 YFGTKQQLFAAAIHLPIDPKLVLKRMQNT----PTAELGFALPSMLLPMW
MAV_3088|M.avium_104 YFGTKEKLFAAAVQIPIDPMQVIGPLREV----PVDDLGYALPSMLLPLW
MAB_2351c|M.abscessus_ATCC_199 YFGTKLDLFREVVQLPVDPSVVLQPLRDV----PVDELGVTIPRLIIALW
MMAR_3397|M.marinum_M YFGSKAELFRSIVGWPFEADAMSALILSG----DRDELGARLTRVFLEAW
TH_3542|M.thermoresistible__bu LFGSKAELFRESLELILDPEMLVAAMT-----GDDGDVGTRMVRVYLQIW
Mvan_0641|M.vanbaalenii_PYR-1 LFGSKEQLFREALRLILDPEVLVAALAGVE--GDSSDVGTRMVRTYLGIW
Mflv_0253|M.gilvum_PYR-GCK LFGSKEALFRESLRLILDPGVLIEALS-----GGEADIGTRMVRAYLGVW
MSMEG_2182|M.smegmatis_MC2_155 FFGNKEGLFNAAVLDTPEHPLHQLAALLD---DGPEEIGARLVREVIRRW
MLBr_02568|M.leprae_Br4923 YYGSKEDLFGACLNREMSRFIAMVRADIDFTQSPKDLLRNTIVSFLRYID
:*.* ** : . : :
Mb1711c|M.bovis_AF2122/97 DS-ELGAGLIATLRSLISGSD-VGLARSFLEEVVTVELGSRVDNPPGTGK
Rv1685c|M.tuberculosis_H37Rv DS-ELGAGLIATLRSLISGSD-VGLARSFLEEVVTVELGSRVDNPPGTGK
MUL_2781|M.ulcerans_Agy99 DS-ELGTGLIASLRSLFGGGD-VDLARSFLQEVVTAEMASLVDNPAGTGM
MAV_3088|M.avium_104 DS-EVGAAFIATLRSILAGEE-ISLFRTFIQDVIGVEVGPRVDNPAGSGV
MAB_2351c|M.abscessus_ATCC_199 DS-ELGANMLAVFRSALTGAD-DGLVRVFFREVLVNIIAERVDSPPGSGV
MMAR_3397|M.marinum_M ERPESRAPLLAILRGAATHEESTTLVRQFFQDQMYPQLAQPLPGP--DTE
TH_3542|M.thermoresistible__bu ESPDTGPTIRAMLQSATSNTDAREAFRSFLREYLLTAVSG-VLGGGEQAR
Mvan_0641|M.vanbaalenii_PYR-1 EAADSAASMRAMLASATSNPDAHEAFRSFMQDYVLTAVSG-VLGGGEQAR
Mflv_0253|M.gilvum_PYR-GCK ESPESAESMRTMLASATSNPDAREAFRGFIQDYVLAAVVG-TLGGDDRAR
MSMEG_2182|M.smegmatis_MC2_155 DEGGSFDLLLTVFKSADIHPLARKLLHDSLAGPVAQRVAA-EFG-VDDAE
MLBr_02568|M.leprae_Br4923 ANRASWIVMYIQAMSLPAFAQTVREARDQITELVAGLMRVGTRSSRPDHE
: . : : : : .
Mb1711c|M.bovis_AF2122/97 IRTQFVASQLMGVVMARYIVRIEPFASLPAEQIVQTIAPNLQRYLTGELP
Rv1685c|M.tuberculosis_H37Rv IRTQFVASQLMGVVMARYIVRIEPFASLPAEQIVQTIAPNLQRYLTGELP
MUL_2781|M.ulcerans_Agy99 IRAQLVASQLLGVVMARYILQIGPFASMPAEDIARAIAPNLQRYLTGEI-
MAV_3088|M.avium_104 IRIQFVASQLVGVVMARYILELEPFASLPPEQIAATIAPNLQRYLTGDLP
MAB_2351c|M.abscessus_ATCC_199 LRAEFAITQMAGILVGRYIMAIEPLASLTAEQIALTVGPNIQRYLTGALP
MMAR_3397|M.marinum_M LRIDLAMAQLLGIAYLRHVLRLEPIASTPLEEVVARVTPTIIAHLEPSRL
TH_3542|M.thermoresistible__bu LRAELAATGLVGTAMLRYIMRVPPLAELSTEEVVRMLAPTVNRYLTADAD
Mvan_0641|M.vanbaalenii_PYR-1 LRAMLAASSLVGTAMLRYVMRVAPLAELPTEDVVRLVAPTVTRYLTADAR
Mflv_0253|M.gilvum_PYR-GCK LRAMLAASSLVGTAMMRYLMEIPPLSELATEDVIRLVAPTVTRYLTAPAD
MSMEG_2182|M.smegmatis_MC2_155 LRVELVTVHMVGLAFARYQLKIEPLASAGLDDLVSWLGPTVQRYLTAPDP
MLBr_02568|M.leprae_Br4923 YEIMAGALVGAGEAMATRLTTGDIAVDEAAELMINLFWHGLKGAPADRDF
. * . : : . :
Mb1711c|M.bovis_AF2122/97 DDLAP-------
Rv1685c|M.tuberculosis_H37Rv DDLAP-------
MUL_2781|M.ulcerans_Agy99 ------------
MAV_3088|M.avium_104 DGLAP-------
MAB_2351c|M.abscessus_ATCC_199 S-ITP-------
MMAR_3397|M.marinum_M DKAADR------
TH_3542|M.thermoresistible__bu ELGLPSSLTAGS
Mvan_0641|M.vanbaalenii_PYR-1 ELGLPD------
Mflv_0253|M.gilvum_PYR-GCK ELGLPG------
MSMEG_2182|M.smegmatis_MC2_155 ------------
MLBr_02568|M.leprae_Br4923 GPSIVAG-----