For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAAPDNSRRRPGRPAGSSDTRERILSSARELFAHNGIDRTSIRAVAAKAGVDAALVHHYFGTKQQLFAAA IHIPIDPMVIIGPIREAPVEELGYKLPSLLLPIWDSELGAGLIATLRSLISGSDVGLARSFLEEVVTVEL GSRVDNPPGTGKIRTQFVASQLMGVVMARYIVRIEPFASLPAEQIVQTIAPNLQRYLTGELPDDLAP
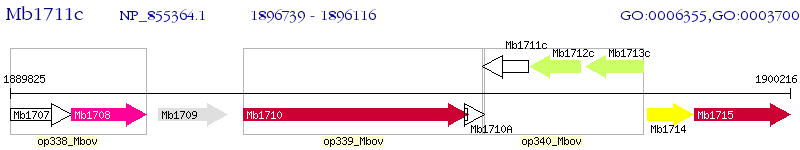
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1711c | - | - | 100% (207) | hypothetical protein Mb1711c |
| M. bovis AF2122 / 97 | Mb0264c | - | 6e-09 | 30.71% (140) | hypothetical protein Mb0264c |
| M. bovis AF2122 / 97 | Mb0149 | - | 3e-08 | 31.58% (76) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0253 | - | 5e-26 | 32.50% (200) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv1685c | - | 1e-114 | 100.00% (207) | hypothetical protein Rv1685c |
| M. leprae Br4923 | MLBr_00316 | - | 3e-06 | 27.60% (192) | putative TetR/AcrR-family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_2351c | - | 2e-62 | 54.59% (196) | TetR family transcriptional regulator |
| M. marinum M | MMAR_2484 | - | 2e-83 | 71.36% (206) | hypothetical protein MMAR_2484 |
| M. avium 104 | MAV_3088 | - | 1e-83 | 71.01% (207) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_3765 | - | 2e-83 | 73.98% (196) | transcriptional regulator |
| M. thermoresistible (build 8) | TH_3542 | - | 4e-24 | 33.52% (179) | PUTATIVE transcriptional regulator, TetR family |
| M. ulcerans Agy99 | MUL_2781 | - | 2e-78 | 73.63% (201) | hypothetical protein MUL_2781 |
| M. vanbaalenii PYR-1 | Mvan_3298 | - | 6e-82 | 72.31% (195) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0253|M.gilvum_PYR-GCK MPPPAQT-----------------------------------------GR
TH_3542|M.thermoresistible__bu --------------------------------------------------
Mb1711c|M.bovis_AF2122/97 MAAPDNS-----------------------------------------RR
Rv1685c|M.tuberculosis_H37Rv MAAPDNS-----------------------------------------RR
MMAR_2484|M.marinum_M METTNTG-----------------------------------------RR
MUL_2781|M.ulcerans_Agy99 MQPTTTHPAQARRLRSSGKLLGARDVSAHNKPRNVDNTRGGNAPANERRR
MSMEG_3765|M.smegmatis_MC2_155 MTATSDTGRA--------------------------------------RK
Mvan_3298|M.vanbaalenii_PYR-1 MTTNAD------------------------------------------RK
MAV_3088|M.avium_104 MTTTGTT-----------------------------------------RR
MAB_2351c|M.abscessus_ATCC_199 MS----------------------------------------------SK
MLBr_00316|M.leprae_Br4923 MSVPRSG-------------------------------------LSVQMA
Mflv_0253|M.gilvum_PYR-GCK RRGRRKG--DPVSRETVLAAAKGRFARDGFEKTTLRAIAEDAGVDASMVL
TH_3542|M.thermoresistible__bu ----------------VLAAAKRRFAQDGYEKTTLRAIARDAHVDASMVL
Mb1711c|M.bovis_AF2122/97 RPGRPAG--SSDTRERILSSARELFAHNGIDRTSIRAVAAKAGVDAALVH
Rv1685c|M.tuberculosis_H37Rv RPGRPAG--SSDTRERILSSARELFAHNGIDRTSIRAVAAKAGVDAALVH
MMAR_2484|M.marinum_M RPGRPTG--TSNTREHILTCARELFALNGLDRTSVRSVAAAAGVDASLVH
MUL_2781|M.ulcerans_Agy99 RPGRLRGN-SSDTRQRILASARELFASNGIDRTSIRAVAAAADVDAALVH
MSMEG_3765|M.smegmatis_MC2_155 RPGRPPG--ASDTRDRILASARELFARNGFDKTSIRAVAAGADVDPALVH
Mvan_3298|M.vanbaalenii_PYR-1 RPGRPPG--TSDTRDRILECARELFARNGITNTSIRSIAAGAGVDPALVH
MAV_3088|M.avium_104 RRGRPAG--SSGSRERILASARELFARNGIRNTSIRAVAAAAGVDSALVH
MAB_2351c|M.abscessus_ATCC_199 RPGRRPG--TSSARDDILASARKLFSLNGIDKTSIRAIASDAGVDPALVH
MLBr_00316|M.leprae_Br4923 QPARPLRADAARNRARILGVAYEAFATEGLS-VPIDEIARRAGVGAGTVY
:* * *: :* ..: :* * *... *
Mflv_0253|M.gilvum_PYR-GCK YLFGSKEALFRESL--RLILDPGVLIEALSGG-EADIGTRMVRAYLGVWE
TH_3542|M.thermoresistible__bu YLFGSKAELFRESL--ELILDPEMLVAAMTGD-DGDVGTRMVRVYLQIWE
Mb1711c|M.bovis_AF2122/97 HYFGTKQQLFAAAI--HIPIDPMVIIGPIREAPVEELGYKLPSLLLPIWD
Rv1685c|M.tuberculosis_H37Rv HYFGTKQQLFAAAI--HIPIDPMVIIGPIREAPVEELGYKLPSLLLPIWD
MMAR_2484|M.marinum_M HYYGTKQQLFAAAI--QIPIDPTVVLEQIVETPVDELGLKLPSVLLPLWD
MUL_2781|M.ulcerans_Agy99 HYFGTKQQLFAAAI--HLPIDPKLVLKRMQNTPTAELGFALPSMLLPMWD
MSMEG_3765|M.smegmatis_MC2_155 HYFGTKTQLFAAAI--HIPIDPLEVIGPLREVPVEQIGHTLPRLLLPLWD
Mvan_3298|M.vanbaalenii_PYR-1 HYFGTKTQLFAAAI--EIPIDPMSIIGPLKEVPVDQLGRALPSLLLPLWD
MAV_3088|M.avium_104 HYFGTKEKLFAAAV--QIPIDPMQVIGPLREVPVDDLGYALPSMLLPLWD
MAB_2351c|M.abscessus_ATCC_199 HYFGTKLDLFREVV--QLPVDPSVVLQPLRDVPVDELGVTIPRLIIALWD
MLBr_00316|M.leprae_Br4923 RHFPTKEALCAAVIGDRMPHLVDDGYVLLKSAGPGEALFTYLRSLVLHWG
: :* * : .: : : : *
Mflv_0253|M.gilvum_PYR-GCK SPESAESMRTMLASATSNPDAREAFRGFIQDYVLAAVVGTLGG-DDRARL
TH_3542|M.thermoresistible__bu SPDTGPTIRAMLQSATSNTDAREAFRSFLREYLLTAVSGVLGG-GEQARL
Mb1711c|M.bovis_AF2122/97 S-ELGAGLIATLRSLISGSDV-GLARSFLEEVVTVELGSRVDNPPGTGKI
Rv1685c|M.tuberculosis_H37Rv S-ELGAGLIATLRSLISGSDV-GLARSFLEEVVTVELGSRVDNPPGTGKI
MMAR_2484|M.marinum_M S-ELGTGLVSTLRALMAGTES-NLARSFLQEVVTSEVGSRVDSPTGTGRI
MUL_2781|M.ulcerans_Agy99 S-ELGTGLIASLRSLFGGGDV-DLARSFLQEVVTAEMASLVDNPAGTGMI
MSMEG_3765|M.smegmatis_MC2_155 S-ELGKGFIATLRSILAGNDV-SLVRSFLQEVIVGEVGSRVDNPAGSGRI
Mvan_3298|M.vanbaalenii_PYR-1 S-ELGKGFIATLRSILAGSDV-SMVRSFLQDLIATEVGGRVDDPPGSGRV
MAV_3088|M.avium_104 S-EVGAAFIATLRSILAGEEI-SLFRTFIQDVIGVEVGPRVDNPAGSGVI
MAB_2351c|M.abscessus_ATCC_199 S-ELGANMLAVFRSALTGADD-GLVRVFFREVLVNIIAERVDSPPGSGVL
MLBr_00316|M.leprae_Br4923 A--TDRGLVDALAGAG-GIDILGVAP-DAEDAFLTILSDLLYAAQYAGTA
: : : . . : .: . : : .
Mflv_0253|M.gilvum_PYR-GCK RAMLAASSLVGTAMMRYLMEIPPLSELATEDVIRLVAPTVTRYLTAPADE
TH_3542|M.thermoresistible__bu RAELAATGLVGTAMLRYIMRVPPLAELSTEEVVRMLAPTVNRYLTADADE
Mb1711c|M.bovis_AF2122/97 RTQFVASQLMGVVMARYIVRIEPFASLPAEQIVQTIAPNLQRYLTGELPD
Rv1685c|M.tuberculosis_H37Rv RTQFVASQLMGVVMARYIVRIEPFASLPAEQIVQTIAPNLQRYLTGELPD
MMAR_2484|M.marinum_M RAQFVASQLLGVVMARHIVKIEPFASLPADQVAQAIAPTLQRYLTGELPE
MUL_2781|M.ulcerans_Agy99 RAQLVASQLLGVVMARYILQIGPFASMPAEDIARAIAPNLQRYLTGEI--
MSMEG_3765|M.smegmatis_MC2_155 RVQYVASQLVGVVMARYILELEPFKSLPVEQIAETIGPNLQRYLTGELPG
Mvan_3298|M.vanbaalenii_PYR-1 RVQFVASQLVGIVMARYILELEPFKSLPVEQIVDTVAPNLQRYLTGELPQ
MAV_3088|M.avium_104 RIQFVASQLVGVVMARYILELEPFASLPPEQIAATIAPNLQRYLTGDLPD
MAB_2351c|M.abscessus_ATCC_199 RAEFAITQMAGILVGRYIMAIEPLASLTAEQIALTVGPNIQRYLTGALPS
MLBr_00316|M.leprae_Br4923 RTDVGVREVKSILVG--CQAMEAYNSALAERVTDVVVDGLRAAR------
* : . : : . . : : : :
Mflv_0253|M.gilvum_PYR-GCK LGLPG------
TH_3542|M.thermoresistible__bu LGLPSSLTAGS
Mb1711c|M.bovis_AF2122/97 DLAP-------
Rv1685c|M.tuberculosis_H37Rv DLAP-------
MMAR_2484|M.marinum_M EIAS-------
MUL_2781|M.ulcerans_Agy99 -----------
MSMEG_3765|M.smegmatis_MC2_155 FGALD------
Mvan_3298|M.vanbaalenii_PYR-1 FSGD-------
MAV_3088|M.avium_104 GLAP-------
MAB_2351c|M.abscessus_ATCC_199 ITP--------
MLBr_00316|M.leprae_Br4923 -----------