For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTRTVVVTGAGSGIGRAIASTLAQRDWRVVVTDVNGEAARSVAAELPNPSAGHESAPLDVTSPDAAAAVA SDIAERFGLDAWVSNAGISFMHRFLDAPIERFDQTMDVNLKGVFVCGQAAAREMVRSGVPGVIVNTASMA GKQGRVPFLADYVASKFGVVGLTQAMAYELGEHGITVNCVCPGFVETPMQSRELEWEAELRGTTPDGVRA MMIADTPLGRLEQADDVARAVAFLLSDDARFITGEALAVNGGAYMD
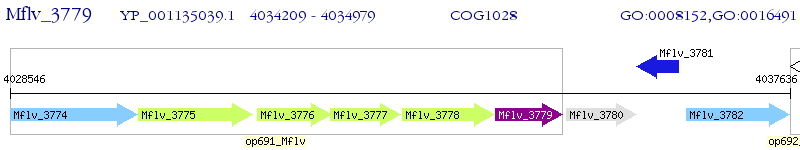
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3779 | - | - | 100% (256) | short-chain dehydrogenase/reductase SDR |
| M. gilvum PYR-GCK | Mflv_2469 | - | 3e-37 | 38.43% (255) | short-chain dehydrogenase/reductase SDR |
| M. gilvum PYR-GCK | Mflv_2501 | - | 1e-36 | 38.89% (252) | short-chain dehydrogenase/reductase SDR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2882c | - | 8e-35 | 37.94% (253) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv2857c | - | 8e-35 | 37.94% (253) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 6e-30 | 33.98% (256) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_4680c | - | 3e-33 | 39.44% (251) | putative short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_2869 | - | 3e-35 | 37.45% (251) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_5088 | fabG | 2e-39 | 39.37% (254) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. smegmatis MC2 155 | MSMEG_3112 | - | 1e-101 | 73.12% (253) | estradiol 17-beta-dehydrogenase 8 |
| M. thermoresistible (build 8) | TH_4254 | - | 6e-36 | 38.25% (251) | PUTATIVE putative 2,5-dichloro-2,5-cyclohexadiene-1,4-diol |
| M. ulcerans Agy99 | MUL_2885 | - | 8e-36 | 37.45% (251) | short-chain type dehydrogenase/reductase |
| M. vanbaalenii PYR-1 | Mvan_2620 | - | 1e-134 | 91.80% (256) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2882c|M.bovis_AF2122/97 --------MMDLS---QRLAGRVAVITGGGSGIGLAAGRRMRAEGATIVV
Rv2857c|M.tuberculosis_H37Rv --------MMDLS---QRLAGRVAVITGGGSGIGLAAGRRMRAEGATIVV
Mflv_3779|M.gilvum_PYR-GCK ---------MTR----------TVVVTGAGSGIGRAIASTLAQRDWRVVV
Mvan_2620|M.vanbaalenii_PYR-1 ---------MTR----------TVVVTGAGSGIGRAIANTLAQRNWRVVV
MSMEG_3112|M.smegmatis_MC2_155 ---------MSTPANSTPDTPSTVVVTGAGSGIGRAIATTLAERGWRVVA
MAB_4680c|M.abscessus_ATCC_199 ---------MNG--NSALLQGKNVIVTGGARGLGAAFSRHIVSQGGRVVI
TH_4254|M.thermoresistible__bu ---------MTA--GSTRLAGKVVLVTGAGGGLGTAIARRLADEGARLVL
MMAR_2869|M.marinum_M ---------MRYP----DLAGKATIVTGAGAGIGLAIAERLAAEGAAVLC
MUL_2885|M.ulcerans_Agy99 ---------MRYP----DLAGKATIVTGAGAGIGLAIAERLAAEGAAVLC
MAV_5088|M.avium_104 ---------MAES----LLLGKVAVVTGAGQGIGREIARALHRHGARVVL
MLBr_01807|M.leprae_Br4923 MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLAVARRLVADGHRVAV
::**. *:* . : . :
Mb2882c|M.bovis_AF2122/97 GDVDVEAGGAAADELSG-----LFVPTDVCDEDAVNGLFDGAAETYGRID
Rv2857c|M.tuberculosis_H37Rv GDVDVEAGGAAADELSG-----LFVPTDVCDEDAVNGLFDGAAETYGRID
Mflv_3779|M.gilvum_PYR-GCK TDVNGEAARSVAAELPNPSAGHESAPLDVTSPDAAAAVASDIAERFG-LD
Mvan_2620|M.vanbaalenii_PYR-1 TDVNAAAASEVAAALPNPPAGHESAQLDVTSPENAAAVASDVSERLG-LD
MSMEG_3112|M.smegmatis_MC2_155 TDVDVAAADETASSLSG--SGHESARLDVTDPAQASALADDVADRIG-LG
MAB_4680c|M.abscessus_ATCC_199 ADVLDGDGRQLAAELGDAARFTH---LDVTDAEQWRELIDTTLAGFGTIT
TH_4254|M.thermoresistible__bu ADLQESQCAALADELPAPAGAHHPLGFDVADETAWISAISTVSAQYGALH
MMAR_2869|M.marinum_M ADIDGASAEQAAAKIG-SGA--VGYQVDVSAEEQVAGMVEACVTSFGGLD
MUL_2885|M.ulcerans_Agy99 ADIDGASAEQAAAKIG-SGA--VGYQVDVSAEEQVAGMVEACVTSFGGLD
MAV_5088|M.avium_104 ADLDGSAARSAAAQIDDSGANCTGLACDVTSEEQVGALVAGTVREHGGLD
MLBr_01807|M.leprae_Br4923 THRASVVP-----------DWFFGVECDVTDNDAIGAAFKAVEEHQGPVE
. ** * :
Mb2882c|M.bovis_AF2122/97 IAFNNAGISPPEDNLIENTELAAWQRVQDVNLKSVYLCCRAALRHMVLAG
Rv2857c|M.tuberculosis_H37Rv IAFNNAGISPPEDNLIENTELAAWQRVQDVNLKSVYLCCRAALRHMVLAG
Mflv_3779|M.gilvum_PYR-GCK AWVSNAGISFMHR--FLDAPIERFDQTMDVNLKGVFVCGQAAAREMVRSG
Mvan_2620|M.vanbaalenii_PYR-1 AWVSNAGISFMHR--FLDAPIERYEQTMDVNLKGVFVCGQAAAREMVRAG
MSMEG_3112|M.smegmatis_MC2_155 AWVSNAGISAMAH--FVDISAEQLDRSLDVNLKGVFFCGQAAARAMIRTG
MAB_4680c|M.abscessus_ATCC_199 GLVNNAGISSAAM--VADEPLEHFRKVIEINLVGVFIGMQAVIPAMRAHG
TH_4254|M.thermoresistible__bu ALVNNAAIGALGN--VAEERLERWDRILDVGATGVWLGMKHAVPLIRRSG
MMAR_2869|M.marinum_M KLVANAGVVHLAP--ILDTAVEDFDRVIGINLRGAWLCTKHAAPKMVERG
MUL_2885|M.ulcerans_Agy99 KLVANAGVVHLAP--ILDTAVEDFDRVIGINLRGAWLCTKHAAPKMVERG
MAV_5088|M.avium_104 VFVNNAGITRDAS--LKRMTVADFDAVIAVHLRGTWLGVREAAAVMRERK
MLBr_01807|M.leprae_Br4923 VLVANAGITSDML--IMRMSEEQFQKVIDVNLTGVFRVAKLASRNMIKQR
. **.: . : ..: : . :
Mb2882c|M.bovis_AF2122/97 K-GSIVNTASFVAVMGSATSQISYTASKGGVLAMSRELGVQFARQGIRVN
Rv2857c|M.tuberculosis_H37Rv K-GSIVNTASFVAVMGSATSQISYTASKGGVLAMSRELGVQFARQGIRVN
Mflv_3779|M.gilvum_PYR-GCK VPGVIVNTASMAGKQGRVPFLADYVASKFGVVGLTQAMAYELGEHGITVN
Mvan_2620|M.vanbaalenii_PYR-1 IPGMIVNTASMAGKQGRVPFLSDYVASKFGVVGLTQAMAYELGEHGITVN
MSMEG_3112|M.smegmatis_MC2_155 VRGTIVNTASMAAKQGRVPFLADYVASKFGVLGLTQAMAFELAAHGITVN
MAB_4680c|M.abscessus_ATCC_199 C-GSIVNISSAAGLVG-LALTSGYGASKWGVRGLSKVAAIELGRERIRVN
TH_4254|M.thermoresistible__bu G-GSVVNICSIFGTVGGFGDSAAYHAAKGAVRTLTKNAALHWAPDGVRVN
MMAR_2869|M.marinum_M G-GAIVNMSSLAGHIG-VGGTAAYGMSKAGISHLSRVTAAELRSANVRSN
MUL_2885|M.ulcerans_Agy99 G-GAIVNMSSLAGHIG-VGGTAAYGMSKAGISHLSRVTAAELRSANVRSN
MAV_5088|M.avium_104 T-GSIVNMSSLSGKAG-NPGQTNYSAAKAGIVGLTKAAAKELAHHNVRVN
MLBr_01807|M.leprae_Br4923 F-GRYIFMGSVVGLSG-VPGQANYAASKSGLIGMARSLARELGSRSITAN
* : * . * * :* .: ::: . . : *
Mb2882c|M.bovis_AF2122/97 ALCPGPVNTPLLQELFAKNPERAAR--------RMVHVPLGRFA----EP
Rv2857c|M.tuberculosis_H37Rv ALCPGPVNTPLLQELFAKNPERAAR--------RMVHVPLGRFA----EP
Mflv_3779|M.gilvum_PYR-GCK CVCPGFVETPMQSRELEWEAELRGTTPDGVRAMMIADTPLGRLE----QA
Mvan_2620|M.vanbaalenii_PYR-1 CVCPGFVETPMQSRELQWEAELRGTTPDGVRAMMIADTPLGRLE----QA
MSMEG_3112|M.smegmatis_MC2_155 SVCPGFVATPMQTRELEWEAKLSGSTPDGVRQSWIDATPLGRLQ----TP
MAB_4680c|M.abscessus_ATCC_199 SVHPGVIYTPMT--AKEGFREGEG---------NMPIAAMGRVG----NA
TH_4254|M.thermoresistible__bu SLHPGFIETPQLRELYKGTERYER---------MLAGTPLGRLG----RP
MMAR_2869|M.marinum_M ALLPAFVDTPMQQSAMAMFDEALG--------TGGANTMIARLQGRMARP
MUL_2885|M.ulcerans_Agy99 ALLPAFVDTPMQQSAMAMFDEVLG--------TGGANTMIARLQGRMARP
MAV_5088|M.avium_104 AIQPGLIRTPMTA---AMPPDVFA--------EREAAVPMKRAG----EP
MLBr_01807|M.leprae_Br4923 VVAPGFIDTEMTRAMDSKYQEAALQ-----------AIPLGRIG----AV
: *. : * : *
Mb2882c|M.bovis_AF2122/97 DEIAAAVAFLASDDASFITASTFLVDGGISSAYVTPL----
Rv2857c|M.tuberculosis_H37Rv DEIAAAVAFLASDDASFITASTFLVDGGISSAYVTPL----
Mflv_3779|M.gilvum_PYR-GCK DDVARAVAFLLSDDARFITGEALAVNGGAYMD---------
Mvan_2620|M.vanbaalenii_PYR-1 DDVARAVAFLLSDDARFITGEALAVNGGAYMD---------
MSMEG_3112|M.smegmatis_MC2_155 DDVARAVAFLVSEDARFITGEALSVNGGAYMD---------
MAB_4680c|M.abscessus_ATCC_199 DEVAGAVAYLLSDAASYVTGAELAVDGGWTAGPTLTTITGQ
TH_4254|M.thermoresistible__bu DEIAAVVAFLVSDDASFITGSEVYADGGWTAR---------
MMAR_2869|M.marinum_M EEMASIVAFLLSDDSSMITGTTQIADGGTIAALW-------
MUL_2885|M.ulcerans_Agy99 EEMARIVAFLLPDDSSMITGTTQIADGGTIAALW-------
MAV_5088|M.avium_104 EEVAGAVVFLASELSSYITGTVLGVGGGRYM----------
MLBr_01807|M.leprae_Br4923 EEIAGAVSFLASEDAGYISGAVIPVDGGLGMGH--------
:::* * :* .: : ::. ..**