For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NGNSALLQGKNVIVTGGARGLGAAFSRHIVSQGGRVVIADVLDGDGRQLAAELGDAARFTHLDVTDAEQW RELIDTTLAGFGTITGLVNNAGISSAAMVADEPLEHFRKVIEINLVGVFIGMQAVIPAMRAHGCGSIVNI SSAAGLVGLALTSGYGASKWGVRGLSKVAAIELGRERIRVNSVHPGVIYTPMTAKEGFREGEGNMPIAAM GRVGNADEVAGAVAYLLSDAASYVTGAELAVDGGWTAGPTLTTITGQ*
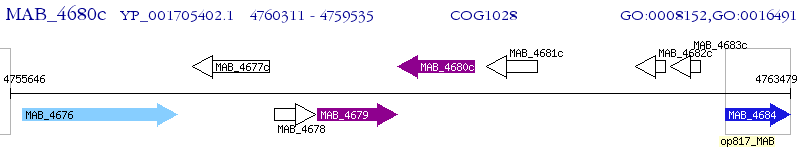
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4680c | - | - | 100% (258) | putative short-chain dehydrogenase/reductase |
| M. abscessus ATCC 19977 | MAB_1219 | - | 8e-31 | 36.03% (247) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_2723c | - | 9e-30 | 33.89% (239) | 3-oxoacyl- |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2025 | fabG3 | 1e-59 | 47.18% (248) | 20-beta-hydroxysteroid dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4505 | - | 1e-57 | 46.12% (245) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv2002 | fabG3 | 1e-59 | 47.18% (248) | 20-beta-hydroxysteroid dehydrogenase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 3e-31 | 34.31% (239) | 3-oxoacyl- |
| M. marinum M | MMAR_2981 | fabG3_1 | 1e-60 | 49.17% (242) | 20-beta-hydroxysteroid dehydrogenase FabG3_1 |
| M. avium 104 | MAV_2509 | - | 3e-60 | 49.17% (242) | 3-alpha-(or 20-beta)-hydroxysteroid dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_3515 | - | 7e-62 | 50.42% (236) | 3-alpha-(or 20-beta)-hydroxysteroid dehydrogenase |
| M. thermoresistible (build 8) | TH_4254 | - | 9e-47 | 40.16% (254) | PUTATIVE putative 2,5-dichloro-2,5-cyclohexadiene-1,4-diol |
| M. ulcerans Agy99 | MUL_1037 | fabG3 | 4e-57 | 47.06% (238) | 20-beta-hydroxysteroid dehydrogenase FabG3 |
| M. vanbaalenii PYR-1 | Mvan_3000 | - | 8e-58 | 47.13% (244) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2025|M.bovis_AF2122/97 ----------MSGR----LIGKVALVSGGARGMGASHVRAMVAEGAKVVF
Rv2002|M.tuberculosis_H37Rv ----------MSGR----LIGKVALVSGGARGMGASHVRAMVAEGAKVVF
MMAR_2981|M.marinum_M ----------MAGR----LTGKVALVSGGARGMGASHVRALVAEGAHVVL
MAV_2509|M.avium_104 ----------MAER----LAGKVALVSGGARGMGASHVRSLVAEGAKVVF
Mflv_4505|M.gilvum_PYR-GCK ----------MTGR----LAGKVALISGAARGMGASHARVMAGHGAKVVC
MSMEG_3515|M.smegmatis_MC2_155 -----------MGR----VAGKVALISGGARGMGAAHARALVAEGAKVVI
Mvan_3000|M.vanbaalenii_PYR-1 -----------MGR----VDGKVALISGGAQGMGAADARALIAEGAKVVI
MUL_1037|M.ulcerans_Agy99 -----------MGR----VDGKVALISGGARGMGASHARLLVQEGAKVVI
MAB_4680c|M.abscessus_ATCC_199 ----------MNGNS-ALLQGKNVIVTGGARGLGAAFSRHIVSQGGRVVI
TH_4254|M.thermoresistible__bu ----------MTAGS-TRLAGKVVLVTGAGGGLGTAIARRLADEGARLVL
MLBr_01807|M.leprae_Br4923 MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLAVARRLVADGHRVAV
. .: :::*. *:* : * : .* ::.
Mb2025|M.bovis_AF2122/97 GDILDEEGKAVAAELA----DAARYVHLDVTQPAQWTAAVDTAVTAFGGL
Rv2002|M.tuberculosis_H37Rv GDILDEEGKAVAAELA----DAARYVHLDVTQPAQWTAAVDTAVTAFGGL
MMAR_2981|M.marinum_M GDILDDEGRAVAAELG----DAARYVHLDVTQPEQWTAAVDTAVNEFGGL
MAV_2509|M.avium_104 GDILDDEGKAVAAEVG----EATRYLHLDVTKPEDWDAAVATALAEFGRI
Mflv_4505|M.gilvum_PYR-GCK GDILDSDGEAVAAELG----DAARYVHLDVTSPDDWDRAVAAAVADFGGL
MSMEG_3515|M.smegmatis_MC2_155 GDILDDEGAALAAELG----DAARYVHLDVTQAEDWDTAVATATTDFGLL
Mvan_3000|M.vanbaalenii_PYR-1 GDILDDKGKALADEINAETPDSIRYVHLDVTQADQWEAAVATAVDAFGKL
MUL_1037|M.ulcerans_Agy99 GDILDEEGKALAEEIG----DAARYVHLDVTQPDQWEAAVATAVDEFGKL
MAB_4680c|M.abscessus_ATCC_199 ADVLDGDGRQLAAELG----DAARFTHLDVTDAEQWRELIDTTLAGFGTI
TH_4254|M.thermoresistible__bu ADLQESQCAALADELPAP-AGAHHPLGFDVADETAWISAISTVSAQYGAL
MLBr_01807|M.leprae_Br4923 THR--------ASVVP----DWFFGVECDVTDNDAIGAAFKAVEEHQGPV
. * : **:. . :. * :
Mb2025|M.bovis_AF2122/97 HVLVNNAGILNIGTIEDYALTEWQRILDVNLTGVFLGIRAVVKPMKEAGR
Rv2002|M.tuberculosis_H37Rv HVLVNNAGILNIGTIEDYALTEWQRILDVNLTGVFLGIRAVVKPMKEAGR
MMAR_2981|M.marinum_M HVLVNNAGILNIGTIEDYALSEWQRILDINVTGVFLGIRAAVKPMKEAGR
MAV_2509|M.avium_104 DVLVNNAGIINIGTLEDYALSEWQRILDINLTGVFLGIRAVVKPMKKAGR
Mflv_4505|M.gilvum_PYR-GCK DVLVNNAGILNIGTVEDYELSEWHRILDVNLTGVFLGIRAATPTMKAAGR
MSMEG_3515|M.smegmatis_MC2_155 NVLINNAGIVALGAIGKFDMTKWQNVIDVNLTGTFLGMQASVGAMKAAGG
Mvan_3000|M.vanbaalenii_PYR-1 NVLVNNAGTVALGQIGQFDMAKWQKVIDVNLTGTFLGMQASVEAMKTAGG
MUL_1037|M.ulcerans_Agy99 DVLVNNVGIVALGQLKKFDLGKWQKVIDVNLTGTFLGMRAAVEPMTAAGS
MAB_4680c|M.abscessus_ATCC_199 TGLVNNAGISSAAMVADEPLEHFRKVIEINLVGVFIGMQAVIPAMRAHGC
TH_4254|M.thermoresistible__bu HALVNNAAIGALGNVAEERLERWDRILDVGATGVWLGMKHAVPLIRRSGG
MLBr_01807|M.leprae_Br4923 EVLVANAGITSDMLIMRMSEEQFQKVIDVNLTGVFRVAKLASRNMIKQRF
*: *.. : .: .::::. .*.: : :
Mb2025|M.bovis_AF2122/97 GSIINISSIEGLAG-TVACHGYTATKFAVRGLTKSTALELGPSGIRVNSI
Rv2002|M.tuberculosis_H37Rv GSIINISSIEGLAG-TVACHGYTATKFAVRGLTKSTALELGPSGIRVNSI
MMAR_2981|M.marinum_M GSIINISSIEGLAG-TIASHGYTVSKFAVRGLTKSTALELGPSGIRVNSI
MAV_2509|M.avium_104 GSIINISSIEGMAG-TIACHGYTATKFAVRGLTKSAALELGPSGIRVNSI
Mflv_4505|M.gilvum_PYR-GCK GSIINIASIEGMAG-TVGCHGYTATKFAVRGLTKSTALELGPFGIRVNSV
MSMEG_3515|M.smegmatis_MC2_155 GSIINVSSIEGLRG-APMVHPYVASKWAVRGLTKSAALELGAHQIRVNSI
Mvan_3000|M.vanbaalenii_PYR-1 GSIINISSIEGLRG-AVMVHPYVASKWAVRGLTKSAALELGSHNIRVNSV
MUL_1037|M.ulcerans_Agy99 GSIINVSSIEGLRG-APAVHPYVASKWAVRGLTKSAALELAPLNIRVNSI
MAB_4680c|M.abscessus_ATCC_199 GSIVNISSAAGLVG-LALTSGYGASKWGVRGLSKVAAIELGRERIRVNSV
TH_4254|M.thermoresistible__bu GSVVNICSIFGTVGGFGDSAAYHAAKGAVRTLTKNAALHWAPDGVRVNSL
MLBr_01807|M.leprae_Br4923 GRYIFMGSVVGLSG-VPGQANYAASKSGLIGMARSLARELGSRSITANVV
* : : * * * * .:* .: ::: * . . : .* :
Mb2025|M.bovis_AF2122/97 HPGLVKTPMTDWV-------PEDIFQTALGRAAEPVEVSNLVVYLASDES
Rv2002|M.tuberculosis_H37Rv HPGLVKTPMTDWV-------PEDIFQTALGRAAEPVEVSNLVVYLASDES
MMAR_2981|M.marinum_M HPGLVKTPMTEWV-------PEDLFQTALGRAAEPMEVSNLVVYLASDES
MAV_2509|M.avium_104 HPGLIKTPMTEWV-------PEDIFQSALGRAAEPKEVSNLVVYLASDES
Mflv_4505|M.gilvum_PYR-GCK HPGLVKTPMADWV-------PEDIFQSALGRIAQPHEVSNLVVYLASDES
MSMEG_3515|M.smegmatis_MC2_155 HPGFIRTPMTEHF-------PDDMLRIPLGRPGEVDEVSSFVVFLASDES
Mvan_3000|M.vanbaalenii_PYR-1 HPGFIRTPMTKHF-------PDNMLRIPLGRPGQPEEVATFVVFLASDES
MUL_1037|M.ulcerans_Agy99 HPGFIRTPMTANL-------PDDMVTIPLGRPAESREVSTFVVFLASDDA
MAB_4680c|M.abscessus_ATCC_199 HPGVIYTPMTAKEGF--REGEGNMPIAAMGRVGNADEVAGAVAYLLSDAA
TH_4254|M.thermoresistible__bu HPGFIETPQLRELYKGTERYERMLAGTPLGRLGRPDEIAAVVAFLVSDDA
MLBr_01807|M.leprae_Br4923 APGFIDTEMTRAMDS--KYQEAALQAIPLGRIGAVEEIAGAVSFLASEDA
**.: * : .:** . *:: * :* *: :
Mb2025|M.bovis_AF2122/97 SYSTGAEFVVDGGTVAGLAHNDFGAVEVSSQPEWVT
Rv2002|M.tuberculosis_H37Rv SYSTGAEFVVDGGTVAGLAHNDFGAVEVSSQPEWVT
MMAR_2981|M.marinum_M SYSTGAEFVVDGGTVAGLAHKDFSAVDVAAQPEWVT
MAV_2509|M.avium_104 SYSTGSEFVVDGGTTAGLGHKDFSNVETDAQPDWVT
Mflv_4505|M.gilvum_PYR-GCK SYSTGAEFVVDGGTLAGLAHKDFSAVDVDQQPEWIT
MSMEG_3515|M.smegmatis_MC2_155 RYATGAEFVMDGGLVNDVPHKL--------------
Mvan_3000|M.vanbaalenii_PYR-1 RYATGAEFVMDGGLTNDVPHK---------------
MUL_1037|M.ulcerans_Agy99 SYATGSEFVMDGGLVTDVPHKQF-------------
MAB_4680c|M.abscessus_ATCC_199 SYVTGAELAVDGGWTAGPTLTTITGQ----------
TH_4254|M.thermoresistible__bu SFITGSEVYADGGWTAR-------------------
MLBr_01807|M.leprae_Br4923 GYISGAVIPVDGGLGMGH------------------
: :*: . ***