For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MDLSQRLAGRVAVITGGGSGIGLAAGRRMRAEGATIVVGDVDVEAGGAAADELSGLFVPTDVCDEDAVNG LFDGAAETYGRIDIAFNNAGISPPEDNLIENTELAAWQRVQDVNLKSVYLCCRAALRHMVLAGKGSIVNT ASFVAVMGSATSQISYTASKGGVLAMSRELGVQFARQGIRVNALCPGPVNTPLLQELFAKNPERAARRMV HVPLGRFAEPDEIAAAVAFLASDDASFITASTFLVDGGISSAYVTPL*
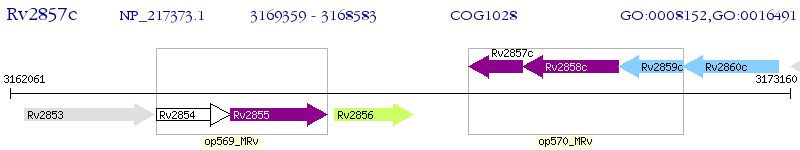
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2857c | - | - | 100% (258) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv2002 | fabG3 | 8e-38 | 38.31% (248) | 20-beta-hydroxysteroid dehydrogenase |
| M. tuberculosis H37Rv | Rv1350 | fabG | 3e-36 | 37.05% (251) | 3-ketoacyl-(acyl-carrier-protein) reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2882c | - | 1e-143 | 100.00% (258) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4062 | - | 1e-110 | 76.65% (257) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 6e-32 | 34.02% (244) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_4680c | - | 3e-30 | 34.01% (247) | putative short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_0387 | fabG3 | 4e-39 | 38.62% (246) | 20-beta-hydroxysteroid dehydrogenase FabG3 |
| M. avium 104 | MAV_3714 | - | 1e-125 | 86.82% (258) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2598 | - | 1e-124 | 85.60% (257) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_4254 | - | 5e-42 | 40.00% (250) | PUTATIVE putative 2,5-dichloro-2,5-cyclohexadiene-1,4-diol |
| M. ulcerans Agy99 | MUL_1037 | fabG3 | 7e-39 | 38.21% (246) | 20-beta-hydroxysteroid dehydrogenase FabG3 |
| M. vanbaalenii PYR-1 | Mvan_2281 | - | 1e-111 | 77.43% (257) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4062|M.gilvum_PYR-GCK ------------MDLSQRLKDKVAVITGGASGIGLATAKRMRSEGAIVVI
Mvan_2281|M.vanbaalenii_PYR-1 ------------MDLGQRLKDKVAVITGGASGIGLATAKRMQAEGAIVVI
Rv2857c|M.tuberculosis_H37Rv -----------MMDLSQRLAGRVAVITGGGSGIGLAAGRRMRAEGATIVV
Mb2882c|M.bovis_AF2122/97 -----------MMDLSQRLAGRVAVITGGGSGIGLAAGRRMRAEGATIVV
MAV_3714|M.avium_104 -----------MMDLTQRLAGRVAVITGAGGGIGLAAARRMHAEGATIVV
MSMEG_2598|M.smegmatis_MC2_155 ------------MDLTQRLAGKVAVITGGASGIGLATGRRLRAEGATVVV
MMAR_0387|M.marinum_M -----------MG----RVDGKVALISGGARGMGASHARLLVQEGAKVVI
MUL_1037|M.ulcerans_Agy99 -----------MG----RVDGKVALISGGARGMGASHARLLVQEGAKVVI
MAB_4680c|M.abscessus_ATCC_199 -----------MNGNSALLQGKNVIVTGGARGLGAAFSRHIVSQGGRVVI
TH_4254|M.thermoresistible__bu -----------MTAGSTRLAGKVVLVTGAGGGLGTAIARRLADEGARLVL
MLBr_01807|M.leprae_Br4923 MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLAVARRLVADGHRVAV
. .: :::*. *:* : .: : :* :.:
Mflv_4062|M.gilvum_PYR-GCK GDLDEKTGKSVANDLN--VTFVQ---VDVSDQVAVDNLFDTAFEVHGGVD
Mvan_2281|M.vanbaalenii_PYR-1 GDVDASTGKSVANDMN--VTFVQ---VDVADQVAVDHLFDTAFEVHGGVD
Rv2857c|M.tuberculosis_H37Rv GDVDVEAGGAAADELS--GLFVP---TDVCDEDAVNGLFDGAAETYGRID
Mb2882c|M.bovis_AF2122/97 GDVDVEAGGAAADELS--GLFVP---TDVCDEDAVNGLFDGAAETYGRID
MAV_3714|M.avium_104 ADIDADAGAAAADELS--GLFVP---TDVADEDAVNALFDTAVRRYGRID
MSMEG_2598|M.smegmatis_MC2_155 GDIDPTTGKAAADELE--GLFVP---VDVSEQEAVDNLFDTAASTFGRVD
MMAR_0387|M.marinum_M GDILDEEGKALAEEIGDAARYVH---LDVTQPDQWEAAVATAVDEFGKLD
MUL_1037|M.ulcerans_Agy99 GDILDEEGKALAEEIGDAARYVH---LDVTQPDQWEAAVATAVDEFGKLD
MAB_4680c|M.abscessus_ATCC_199 ADVLDGDGRQLAAELGDAARFTH---LDVTDAEQWRELIDTTLAGFGTIT
TH_4254|M.thermoresistible__bu ADLQESQCAALADELPAPAGAHHPLGFDVADETAWISAISTVSAQYGALH
MLBr_01807|M.leprae_Br4923 THRASVVPDWFFGVEC-----------DVTDNDAIGAAFKAVEEHQGPVE
. ** : . . * :
Mflv_4062|M.gilvum_PYR-GCK IAFNNAGISPPEDDLIENTGIDAWDRVQDINLKSVFFCCKAALRHMVPAQ
Mvan_2281|M.vanbaalenii_PYR-1 IAFNNAGISPPEDDLIENTGVDAWDRVQDINLKSVFFCCKAALRHMVPAQ
Rv2857c|M.tuberculosis_H37Rv IAFNNAGISPPEDNLIENTELAAWQRVQDVNLKSVYLCCRAALRHMVLAG
Mb2882c|M.bovis_AF2122/97 IAFNNAGISPPEDNLIENTELAAWQRVQDVNLKSVYLCCRAALRHMVLAG
MAV_3714|M.avium_104 IAFNNAGISPPDDDVIENTEPPAWQRVQDVNLKSVYLCCRAALRHMVSAR
MSMEG_2598|M.smegmatis_MC2_155 IAFNNAGISPPEDDLIENTDLPAWQRVQDINLKSVYLSCRAALRHMVPAG
MMAR_0387|M.marinum_M VLVNNAGIVALG--QLKKFDLGKWQKVIDVNLTGTFLGMRAAVEPMTAAG
MUL_1037|M.ulcerans_Agy99 VLVNNVGIVALG--QLKKFDLGKWQKVIDVNLTGTFLGMRAAVEPMTAAG
MAB_4680c|M.abscessus_ATCC_199 GLVNNAGISSAA--MVADEPLEHFRKVIEINLVGVFIGMQAVIPAMRAHG
TH_4254|M.thermoresistible__bu ALVNNAAIGALG--NVAEERLERWDRILDVGATGVWLGMKHAVPLIRRSG
MLBr_01807|M.leprae_Br4923 VLVANAGITSDM--LIMRMSEEQFQKVIDVNLTGVFRVAKLASRNMIKQR
. *..* . : : :: ::. ..: : . :
Mflv_4062|M.gilvum_PYR-GCK KGSIVNTASFVAVNGSATSQISYTASKGGVLAMSRELGIQYARQGIRVNA
Mvan_2281|M.vanbaalenii_PYR-1 KGSIINTASFVAVMGSATSQISYTASKGGVLAMSRELGVQYARQGIRVNA
Rv2857c|M.tuberculosis_H37Rv KGSIVNTASFVAVMGSATSQISYTASKGGVLAMSRELGVQFARQGIRVNA
Mb2882c|M.bovis_AF2122/97 KGSIVNTASFVAVMGSATSQISYTASKGGVLAMSRELGVQFARQGIRVNA
MAV_3714|M.avium_104 RGSIINTASFVAVMGSATSQISYTAAKGGVLALSRELGVQFARQGIRVNA
MSMEG_2598|M.smegmatis_MC2_155 KGSIINTASFVAVMGSATSQISYTASKGGVLAMSRELGVQYARQGIRVNA
MMAR_0387|M.marinum_M SGSIINVSSIEGLRG-APAVHPYVASKWAVRGLTKSAALELAPLNIRVNS
MUL_1037|M.ulcerans_Agy99 SGSIINVSSIEGLRG-APAVHPYVASKWAVRGLTKSAALELAPLNIRVNS
MAB_4680c|M.abscessus_ATCC_199 CGSIVNISSAAGLVG-LALTSGYGASKWGVRGLSKVAAIELGRERIRVNS
TH_4254|M.thermoresistible__bu GGSVVNICSIFGTVGGFGDSAAYHAAKGAVRTLTKNAALHWAPDGVRVNS
MLBr_01807|M.leprae_Br4923 FGRYIFMGSVVGLSG-VPGQANYAASKSGLIGMARSLARELGSRSITANV
* : * . * * *:* .: ::: . . . : .*
Mflv_4062|M.gilvum_PYR-GCK LCPGPVNTPLLQELFAKDPERAARRLVHVPMGRFAEPEELAAAVAFLASD
Mvan_2281|M.vanbaalenii_PYR-1 LCPGPVNTPLLQELFAKDPERAARRLVHVPVGRFAEPEELAAAVAFLASD
Rv2857c|M.tuberculosis_H37Rv LCPGPVNTPLLQELFAKNPERAARRMVHVPLGRFAEPDEIAAAVAFLASD
Mb2882c|M.bovis_AF2122/97 LCPGPVNTPLLQELFAKNPERAARRMVHVPLGRFAEPDEIAAAVAFLASD
MAV_3714|M.avium_104 LCPGPVNTPLLQELFAKDPQRAARRLVHVPVGRFAEPGEIAAAAAFLASD
MSMEG_2598|M.smegmatis_MC2_155 LCPGPVNTPLLQELFAKDPERAARRLVHIPLGRFAEPEELAAAVAFLASD
MMAR_0387|M.marinum_M IHPGFIRTPMTANL--------PDDMVTIPLGRPAESREVSTFVVFLASD
MUL_1037|M.ulcerans_Agy99 IHPGFIRTPMTANL--------PDDMVTIPLGRPAESREVSTFVVFLASD
MAB_4680c|M.abscessus_ATCC_199 VHPGVIYTPMTAKEGF---REGEGNMPIAAMGRVGNADEVAGAVAYLLSD
TH_4254|M.thermoresistible__bu LHPGFIETPQLRELYKGT-ERYERMLAGTPLGRLGRPDEIAAVVAFLVSD
MLBr_01807|M.leprae_Br4923 VAPGFIDTEMTRAMDS---KYQEAALQAIPLGRIGAVEEIAGAVSFLASE
: ** : * : .:** . *:: . :* *:
Mflv_4062|M.gilvum_PYR-GCK DASFITGSSFLVDGGITGHYVTPL----
Mvan_2281|M.vanbaalenii_PYR-1 DASFITASSFLVDGGISGHYVTPL----
Rv2857c|M.tuberculosis_H37Rv DASFITASTFLVDGGISSAYVTPL----
Mb2882c|M.bovis_AF2122/97 DASFITASTFLVDGGISSAYVTPL----
MAV_3714|M.avium_104 DASFITASTFLVDGGISAAYVTPL----
MSMEG_2598|M.smegmatis_MC2_155 DASFITGSTFLVDGGISSAYVTPL----
MMAR_0387|M.marinum_M DASYATGSEFVMDGGLVTDVPHKQF---
MUL_1037|M.ulcerans_Agy99 DASYATGSEFVMDGGLVTDVPHKQF---
MAB_4680c|M.abscessus_ATCC_199 AASYVTGAELAVDGGWTAGPTLTTITGQ
TH_4254|M.thermoresistible__bu DASFITGSEVYADGGWTAR---------
MLBr_01807|M.leprae_Br4923 DAGYISGAVIPVDGGLGMGH--------
*.: :.: . ***