For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VKIQGSVAFVTGANRGLGRQFAEQLLARGAKVYAGARNPETVTLDGVIPVRIDITDPASVQAAAELAADP TILVNNAGTTSYNGLLDGPMDDIRAQMESHFFGTLNVTRAFVPHLVANAPGAILNIASILSWKHIPGIGA YAAAKAALWAQTDAVREELAPKGVAVTGLYVGFMDTDMAAGVDAEKSDPAHIAAAALDGVEAGVAEVLGD EQTRAVKASLSA
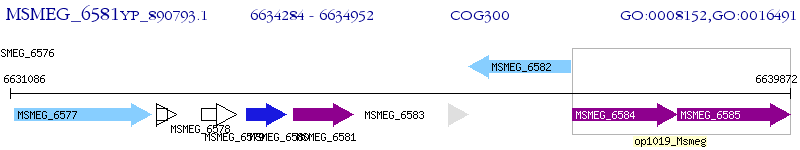
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6581 | - | - | 100% (222) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5711 | - | 2e-71 | 62.04% (216) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_1968 | - | 2e-55 | 48.21% (224) | putative oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3199 | - | 9e-51 | 47.47% (217) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0295 | - | 4e-25 | 35.35% (215) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv3174 | - | 3e-50 | 47.44% (215) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01094 | - | 7e-19 | 29.66% (236) | short chain alcohol dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4428 | - | 1e-39 | 40.91% (220) | short chain dehydrogenase |
| M. marinum M | MMAR_1614 | - | 5e-24 | 38.04% (184) | short chain dehydrogenase |
| M. avium 104 | MAV_2098 | - | 5e-22 | 30.59% (219) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_1828 | - | 2e-22 | 33.47% (236) | PROBABLE SHORT-CHAIN TYPE DEHYDROGENASE/REDUCTASE |
| M. ulcerans Agy99 | MUL_3423 | - | 7e-24 | 37.50% (184) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0234 | - | 7e-73 | 61.47% (218) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_01094|M.leprae_Br4923 MEGFAGKVAVVTGAGSGIGQALAIELARSGAKLAISDVDGEGLAQTEGQL
TH_1828|M.thermoresistible__bu MQGFAGKVAVITGAGSGIGQALALELGRSGAKVAISDVNTEGLTETEDRL
MAV_2098|M.avium_104 -MTHRSPVVLVTGASSGIGRAVAKAFAAKGFEVFGTTRNPRTAEPIPG--
MSMEG_6581|M.smegmatis_MC2_155 -MKIQGSVAFVTGANRGLGRQFAEQLLARG-AKVYAGARNPETVTLDG--
Mvan_0234|M.vanbaalenii_PYR-1 MTDTEHTTALVTGANRGLGRRFAEALVARG-AKVYAAARRPETIDIPG--
Mb3199|M.bovis_AF2122/97 MTSLAERTALVTGANRGMGREYVAQLLGRKVAKVYAATRNPLAIDVSDP-
Rv3174|M.tuberculosis_H37Rv MTSLAERTVLVTGANRGMGREYVAQLLGRKVAKVYAATRNPLAIDVSDP-
MAB_4428|M.abscessus_ATCC_1997 --MIKDTVVLVTGGRRGLGAAIVDEVLNRGARKVYSTARAP--FEDPRA-
MMAR_1614|M.marinum_M ----MS-TWLITGCSTGLGRALAEAVIDAGHHTVATARSVGGVADLVQRS
MUL_3423|M.ulcerans_Agy99 ----MS-TWLITGCSTGLGRALAEAVIDAGHHTVATARSVGGVADLAQRS
Mflv_0295|M.gilvum_PYR-GCK ----MSKVLLISGASRGLGRAITESALAAGHLVVAGVRATSSLSDLATRE
. .::* *:* .
MLBr_01094|M.leprae_Br4923 KAIGASARTDRLDVTEREAFLTYADVVHENFGKVNQIYNNAGIAFTGDVE
TH_1828|M.thermoresistible__bu KAIGAPVKADRLDVTERAAFEAYADAVVEHFGVVNQIYNNAGIAYTGDIE
MAV_2098|M.avium_104 ------VELVELDVTDAASVDAAAKSVIERTGRIDILVNNAGSGILGAAE
MSMEG_6581|M.smegmatis_MC2_155 ------VIPVRIDITDP----ASVQAAAELAADPTILVNNAGTTSYNGLL
Mvan_0234|M.vanbaalenii_PYR-1 ------VVPIQLDITDP----ESVRRAAEQATDVTVVINNAGVSTRAGLL
Mb3199|M.bovis_AF2122/97 -----RVIPLQLDVTDA----VSVAEAADLATDVGILINNAGISRASSVL
Rv3174|M.tuberculosis_H37Rv -----RVIPLQLDVTDA----VSVAEAADLATDVGILINNAGISRASSVL
MAB_4428|M.abscessus_ATCC_1997 -----QVVTKQLEVTSA----ESVDALARDLTDVEIVVNNAGVLHPDPLL
MMAR_1614|M.marinum_M PE---RVLPLALDITEPDQITAAVQAAQQRFGGIDVLVNNAGYGYRAAVE
MUL_3423|M.ulcerans_Agy99 PE---RVLPLALDITEPDQITAAMQAAQQRFGGIDVLVNNAGYGYRAAVE
Mflv_0295|M.gilvum_PYR-GCK PD---RLAVVELDVTDDRHARAAVATAVERFGRLDVLVNNAGYANMSAIE
:::*. : ****
MLBr_01094|M.leprae_Br4923 VSHFKDIERVMDVDYWGVVNGTKAFLPYLISSGDGHVINISSVFGLFSVP
TH_1828|M.thermoresistible__bu VSHFKDIERVMDVDFWGVVNGTKVFLPHLIASGDGHVINISSVFGLFSVP
MAV_2098|M.avium_104 ERSVAQAQRLIDTNFLGLVRLTNAVLPYLRAQGGGRVINIGSVLGFLPQP
MSMEG_6581|M.smegmatis_MC2_155 DGPMDDIRAQMESHFFGTLNVTRAFVPHLVANAPGAILNIASILSWKHIP
Mvan_0234|M.vanbaalenii_PYR-1 DGTLEDIRLEMDTHYFGTLAVTRAFVPVIERNGGGAILNVLSVLSWLHPP
Mb3199|M.bovis_AF2122/97 DKDTSALRGELETNLFGPLALASAFADRIAERS-GAIVNVSSVLAWLPLG
Rv3174|M.tuberculosis_H37Rv DKDTSALRGELETNLFGPLALASAFADRIAERS-GAIVNVSSVLAWLPLG
MAB_4428|M.abscessus_ATCC_1997 TGDFDRIDATFQTNVFGPLRIARAFAPILKANGGGAIVNIHSVLSWL--G
MMAR_1614|M.marinum_M EGDDAEVRDLFETHFFGTVALIKAVLPDMRARRSGAIVNISSIAVALTPV
MUL_3423|M.ulcerans_Agy99 EGDDAEVRDLFETHFFGTVALIKAVLPDMRARRSGAIVNISSIAVALTPV
Mflv_0295|M.gilvum_PYR-GCK DVDVDDFRAQIETNFFGVVNLTRAALPAMRRQRDGHIVQISSVGGRLVRP
:: . * : . : * :::: *:
MLBr_01094|M.leprae_Br4923 GQAAYNSAKFAVRGFTEALREEMALAGRPVNVTTVYPGGIKTAIARNATA
TH_1828|M.thermoresistible__bu GQAAYNAAKFAVRGFTEALRQEMILAGHPVKVTTVHPGGIKTGIARNMTA
MAV_2098|M.avium_104 YGALYAGSKHAVEGYSESLDHETREFG--VRVSVVEPAYTRTSFEANMAD
MSMEG_6581|M.smegmatis_MC2_155 GIGAYAAAKAALWAQTDAVREELAPKG--VAVTGLYVGFMDTDMAA----
Mvan_0234|M.vanbaalenii_PYR-1 TSGAYSAAKAAGWAMTDALRAELAPRG--IHVAALHVGFMDTDMVS----
Mb3199|M.bovis_AF2122/97 MS--YGVSKAAMWSATESMRIELAPRG--VQVVGVYVGLVDTDMGR----
Rv3174|M.tuberculosis_H37Rv MS--YGVSKAAMWSATESMRIELAPRG--VQVVGVYVGLVDTDMGR----
MAB_4428|M.abscessus_ATCC_1997 GAGAYGASKAAIWSVSNTLRLELEAQH--THVLGVHAAFIDTDMVS----
MMAR_1614|M.marinum_M GSGYYAAAKAAMEGMSGALHGELAPLG--ISVTVVEPGAFRTDFAGRSLV
MUL_3423|M.ulcerans_Agy99 GSGYYAAAKAAMEGMSGALHGELAPLG--ISVTVVEPGAFRTDFAGRSLV
Mflv_0295|M.gilvum_PYR-GCK GLAAYQSAKWAVTGFSGVLAVEVAPLG--IKVTVLEPGGMRTDWAGSSMR
* :* * . : : * * : . *
MLBr_01094|M.leprae_Br4923 AEG--------LDVSKIASRFDTWVAHTS--PQHAARIILKA--VRKKKA
TH_1828|M.thermoresistible__bu AEG--------LDAQELARAFDTKLARTS--PEKAAQIILDG--VRKNKA
MAV_2098|M.avium_104 ADAPI---DAYAASREHARRRMAELMKTADDPAIVAEVVLKAATARKPKL
MSMEG_6581|M.smegmatis_MC2_155 ----------------------GVDAE-KSDPAHIAAAALDGVEAGVAE-
Mvan_0234|M.vanbaalenii_PYR-1 ----------------------YIPADQKTDPAVVAELALDGLFAGQPE-
Mb3199|M.bovis_AF2122/97 ----------------------FADAP-KSDPADVVRQVLDGIEAGKED-
Rv3174|M.tuberculosis_H37Rv ----------------------FADAP-KSDPADVVRQVLDGIEAGKED-
MAB_4428|M.abscessus_ATCC_1997 ----------------------ALPVP-KTPPSAIAARIIDALDKGDNE-
MMAR_1614|M.marinum_M QSTRMINDYAATAGQRRKEND-TMHGNQPGDPAKAAAAIITAVESPAPPA
MUL_3423|M.ulcerans_Agy99 QSTRMINDYAATAGQRRKEND-TMHGNQPGDPAKAAAAIITAVESPAPPA
Mflv_0295|M.gilvum_PYR-GCK IAP-VREEYAATVGAAASLSDPNVLG--ASDPAKVAGLLLDVIGMPDPPR
* . :
MLBr_01094|M.leprae_Br4923 RVLVGPDAKVANVVVRFSGGAGYQRLFAQVASRLILNQR-----------
TH_1828|M.thermoresistible__bu RVLVGTDAKILDAIVRLTG-SGYQRLFPKFVARTMPR-------------
MAV_2098|M.avium_104 RYPAGPLARRLSLLKKFAPEAVLDRGIRKTNGLPAASRPHSHLQL-----
MSMEG_6581|M.smegmatis_MC2_155 --VLGDEQTRAVKASLSA--------------------------------
Mvan_0234|M.vanbaalenii_PYR-1 --ILADELTRNVKAQLSAAPA-----------------------------
Mb3199|M.bovis_AF2122/97 --VLADEMSRQVRASLNVPARERIARLMGN--------------------
Rv3174|M.tuberculosis_H37Rv --VLADEMSRQVRASLNVPARERIARLMGN--------------------
MAB_4428|M.abscessus_ATCC_1997 --VLTDDLTVQIKSQLAGPVEKLAYRPGH---------------------
MMAR_1614|M.marinum_M FLLLGPDALMAYRHIAQRRDVEITD-WEQL---TAGTNLTP---------
MUL_3423|M.ulcerans_Agy99 FLLLGPDALMAYRHIAQRRDVEITD-WEQL---TAGTNLTP---------
Mflv_0295|M.gilvum_PYR-GCK RLLVGPDAYR-YATAAGRDLLAVDERWRTLSMSTAADDVSPEQLDPLGSA
MLBr_01094|M.leprae_Br4923 --
TH_1828|M.thermoresistible__bu --
MAV_2098|M.avium_104 --
MSMEG_6581|M.smegmatis_MC2_155 --
Mvan_0234|M.vanbaalenii_PYR-1 --
Mb3199|M.bovis_AF2122/97 --
Rv3174|M.tuberculosis_H37Rv --
MAB_4428|M.abscessus_ATCC_1997 --
MMAR_1614|M.marinum_M --
MUL_3423|M.ulcerans_Agy99 --
Mflv_0295|M.gilvum_PYR-GCK SR