For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TDTEHTTALVTGANRGLGRRFAEALVARGAKVYAAARRPETIDIPGVVPIQLDITDPESVRRAAEQATDV TVVINNAGVSTRAGLLDGTLEDIRLEMDTHYFGTLAVTRAFVPVIERNGGGAILNVLSVLSWLHPPTSGA YSAAKAAGWAMTDALRAELAPRGIHVAALHVGFMDTDMVSYIPADQKTDPAVVAELALDGLFAGQPEILA DELTRNVKAQLSAAPA*
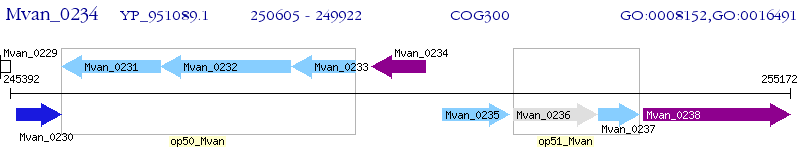
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0234 | - | - | 100% (227) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_1493 | - | 5e-51 | 48.90% (227) | short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_0982 | - | 1e-20 | 34.27% (178) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3199 | - | 1e-51 | 48.67% (226) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0295 | - | 4e-22 | 34.64% (179) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv3174 | - | 4e-51 | 48.23% (226) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_00862 | ephD | 2e-17 | 32.49% (197) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4428 | - | 5e-45 | 45.41% (218) | short chain dehydrogenase |
| M. marinum M | MMAR_1614 | - | 1e-26 | 37.91% (182) | short chain dehydrogenase |
| M. avium 104 | MAV_2098 | - | 7e-24 | 29.96% (247) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5711 | - | 1e-95 | 77.27% (220) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_3670 | - | 1e-20 | 35.39% (178) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_3423 | - | 9e-27 | 37.91% (182) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_0234|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5711|M.smegmatis_MC2_155 --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
MAB_4428|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_1614|M.marinum_M --------------------------------------------------
MUL_3423|M.ulcerans_Agy99 --------------------------------------------------
Mflv_0295|M.gilvum_PYR-GCK --------------------------------------------------
TH_3670|M.thermoresistible__bu --------------------------------------------------
MAV_2098|M.avium_104 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 MPATQSIHKVPHQFVESPDGVRLAIYQSGQPEGPAIVLVHGFPDSHVLWD
Mvan_0234|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5711|M.smegmatis_MC2_155 --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
MAB_4428|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_1614|M.marinum_M --------------------------------------------------
MUL_3423|M.ulcerans_Agy99 --------------------------------------------------
Mflv_0295|M.gilvum_PYR-GCK --------------------------------------------------
TH_3670|M.thermoresistible__bu --------------------------------------------------
MAV_2098|M.avium_104 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 GVVPLLAKRFRIVRYDNRGVGQSSVPKPVSAYTMTRFADDFAAVIDELSP
Mvan_0234|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5711|M.smegmatis_MC2_155 --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
MAB_4428|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_1614|M.marinum_M --------------------------------------------------
MUL_3423|M.ulcerans_Agy99 --------------------------------------------------
Mflv_0295|M.gilvum_PYR-GCK --------------------------------------------------
TH_3670|M.thermoresistible__bu --------------------------------------------------
MAV_2098|M.avium_104 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DRPVHVLAHDWGSVGVWEYLSRSGASDRVASFTSVSGPSQDQLVDYILSG
Mvan_0234|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5711|M.smegmatis_MC2_155 --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
MAB_4428|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_1614|M.marinum_M --------------------------------------------------
MUL_3423|M.ulcerans_Agy99 --------------------------------------------------
Mflv_0295|M.gilvum_PYR-GCK --------------------------------------------------
TH_3670|M.thermoresistible__bu --------------------------------------------------
MAV_2098|M.avium_104 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LRRPWRPRTFARAVSQLFRLTYMVLFSIPVLVPMVLWIALSSATIRRTMV
Mvan_0234|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5711|M.smegmatis_MC2_155 --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
MAB_4428|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_1614|M.marinum_M --------------------------------------------------
MUL_3423|M.ulcerans_Agy99 --------------------------------------------------
Mflv_0295|M.gilvum_PYR-GCK --------------------------------------------------
TH_3670|M.thermoresistible__bu --------------------------------------------------
MAV_2098|M.avium_104 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DNISTDQIHHSATLARDAAHSVKTYPANYFRAFSGRRRGKGVPVVNVPVQ
Mvan_0234|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5711|M.smegmatis_MC2_155 --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
MAB_4428|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_1614|M.marinum_M --------------------------------------------------
MUL_3423|M.ulcerans_Agy99 --------------------------------------------------
Mflv_0295|M.gilvum_PYR-GCK --------------------------------------------------
TH_3670|M.thermoresistible__bu --------------------------------------------------
MAV_2098|M.avium_104 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LIVNTMDRYVRPYGYDATARWVPRLWRRDIKAGHFSPMSHPQVMAAAVHD
Mvan_0234|M.vanbaalenii_PYR-1 ----------------------MTDTEHTTALVTGANRGLGRRFAEALVA
MSMEG_5711|M.smegmatis_MC2_155 ------------------------MTHDIVALVTGANRGLGRRLAAELVA
Mb3199|M.bovis_AF2122/97 ----------------------MTSLAERTALVTGANRGMGREYVAQLLG
Rv3174|M.tuberculosis_H37Rv ----------------------MTSLAERTVLVTGANRGMGREYVAQLLG
MAB_4428|M.abscessus_ATCC_1997 ------------------------MIKDTVVLVTGGRRGLGAAIVDEVLN
MMAR_1614|M.marinum_M --------------------------MS-TWLITGCSTGLGRALAEAVID
MUL_3423|M.ulcerans_Agy99 --------------------------MS-TWLITGCSTGLGRALAEAVID
Mflv_0295|M.gilvum_PYR-GCK --------------------------MSKVLLISGASRGLGRAITESALA
TH_3670|M.thermoresistible__bu --------------------MGSNVWGSKVWFITGASRGFGREWAIAALD
MAV_2098|M.avium_104 -----------------------MTHRSPVVLVTGASSGIGRAVAKAFAA
MLBr_00862|M.leprae_Br4923 LADLVDGKQPSRALLRAQVGRSRDTFGDTLVSVTGAGSGIGRETALALAR
::* *:* .
Mvan_0234|M.vanbaalenii_PYR-1 RG-AKVYAAARRPETIDI-----PG---VVPIQLDITDP----ESVRRAA
MSMEG_5711|M.smegmatis_MC2_155 RG-AKVYAAARRPETLDL-----PG---VVPIPLDITDA----ESVRRAA
Mb3199|M.bovis_AF2122/97 RKVAKVYAATRNPLAIDVSD---PR---VIPLQLDVTDA----VSVAEAA
Rv3174|M.tuberculosis_H37Rv RKVAKVYAATRNPLAIDVSD---PR---VIPLQLDVTDA----VSVAEAA
MAB_4428|M.abscessus_ATCC_1997 RGARKVYSTARAPFEDPR-----AQ---VVTKQLEVTSA----ESVDALA
MMAR_1614|M.marinum_M AGHHTVATARSVGGVADLVQRSPER---VLPLALDITEPDQITAAVQAAQ
MUL_3423|M.ulcerans_Agy99 AGHHTVATARSVGGVADLAQRSPER---VLPLALDITEPDQITAAMQAAQ
Mflv_0295|M.gilvum_PYR-GCK AGHLVVAGVRATSSLSDLATREPDR---LAVVELDVTDDRHARAAVATAV
TH_3670|M.thermoresistible__bu RGDRVAATARDVSTLDDLRDRFGDA---VLAMTLDVTDRAADFAAVQRAH
MAV_2098|M.avium_104 KGFEVFGTTRNPRTAEPIPG--------VELVELDVTDAASVDAAAKSVI
MLBr_00862|M.leprae_Br4923 RGAEVVVSDIDEAAAKNTAAQIVASGGVAYAYALDVSDAAAVEAFAEQVS
*:::.
Mvan_0234|M.vanbaalenii_PYR-1 EQATDVTVVINNAGVSTRAGLLDGTLEDIRLEMDTHYFGTLAVTRAFVP-
MSMEG_5711|M.smegmatis_MC2_155 EIAGDITVLVNNAGISTRANLLTGPVEDIRAEMETHYFGTLNVTRAFVP-
Mb3199|M.bovis_AF2122/97 DLATDVGILINNAGISRASSVLDKDTSALRGELETNLFGPLALASAFAD-
Rv3174|M.tuberculosis_H37Rv DLATDVGILINNAGISRASSVLDKDTSALRGELETNLFGPLALASAFAD-
MAB_4428|M.abscessus_ATCC_1997 RDLTDVEIVVNNAGVLHPDPLLTGDFDRIDATFQTNVFGPLRIARAFAP-
MMAR_1614|M.marinum_M QRFGGIDVLVNNAGYGYRAAVEEGDDAEVRDLFETHFFGTVALIKAVLP-
MUL_3423|M.ulcerans_Agy99 QRFGGIDVLVNNAGYGYRAAVEEGDDAEVRDLFETHFFGTVALIKAVLP-
Mflv_0295|M.gilvum_PYR-GCK ERFGRLDVLVNNAGYANMSAIEDVDVDDFRAQIETNFFGVVNLTRAALP-
TH_3670|M.thermoresistible__bu DHFGRLDIVVNNAGYGHFGFVEELTETEVRDQMETNFFGALWVTQAALP-
MAV_2098|M.avium_104 ERTGRIDILVNNAGSGILGAAEERSVAQAQRLIDTNFLGLVRLTNAVLP-
MLBr_00862|M.leprae_Br4923 AKHGVPDIVVNNAGIGQAGRFLDTPPEEFDRVLAVNLGGVVNCCRAFGQR
:::**** : .: * : *
Mvan_0234|M.vanbaalenii_PYR-1 VIERNGGGAILNVLSVLSWLHPPTSGAYSAAKAAGWAMTDALRAELAPRG
MSMEG_5711|M.smegmatis_MC2_155 NIERNGGGAILNVLSVLSWMHPPTSGAYASAKAAAWALTDAVRTELAPRG
Mb3199|M.bovis_AF2122/97 RIAERSG-AIVNVSSVLAWLPLGMS--YGVSKAAMWSATESMRIELAPRG
Rv3174|M.tuberculosis_H37Rv RIAERSG-AIVNVSSVLAWLPLGMS--YGVSKAAMWSATESMRIELAPRG
MAB_4428|M.abscessus_ATCC_1997 ILKANGGGAIVNIHSVLSWLGGAGA--YGASKAAIWSVSNTLRLELEAQH
MMAR_1614|M.marinum_M DMRARRSGAIVNISSIAVALTPVGSGYYAAAKAAMEGMSGALHGELAPLG
MUL_3423|M.ulcerans_Agy99 DMRARRSGAIVNISSIAVALTPVGSGYYAAAKAAMEGMSGALHGELAPLG
Mflv_0295|M.gilvum_PYR-GCK AMRRQRDGHIVQISSVGGRLVRPGLAAYQSAKWAVTGFSGVLAVEVAPLG
TH_3670|M.thermoresistible__bu YLRRQRSGHIIQVSSIGGVSAFADLGAYHASKWALEGLSQSLSLEVAPFG
MAV_2098|M.avium_104 YLRAQGGGRVINIGSVLGFLPQPYGALYAGSKHAVEGYSESLDHETREFG
MLBr_00862|M.leprae_Br4923 LVERGTGGHIVNVSSMAAYAPLQSLSAYCTSKAATYMFSDCLRAELDAVG
: . :::: *: * :* * : : *
Mvan_0234|M.vanbaalenii_PYR-1 IHVAALHVGFMDTDMVSY--------------------------IPADQK
MSMEG_5711|M.smegmatis_MC2_155 IHVAALHVGYMDTDMVPY--------------------------VPADQK
Mb3199|M.bovis_AF2122/97 VQVVGVYVGLVDTDMGRF--------------------------ADAP-K
Rv3174|M.tuberculosis_H37Rv VQVVGVYVGLVDTDMGRF--------------------------ADAP-K
MAB_4428|M.abscessus_ATCC_1997 THVLGVHAAFIDTDMVSA--------------------------LPVP-K
MMAR_1614|M.marinum_M ISVTVVEPGAFRTDFAGRSLVQSTRMINDYAATAGQRRKEND-TMHGNQP
MUL_3423|M.ulcerans_Agy99 ISVTVVEPGAFRTDFAGRSLVQSTRMINDYAATAGQRRKEND-TMHGNQP
Mflv_0295|M.gilvum_PYR-GCK IKVTVLEPGGMRTDWAGSSMRIAP-VREEYAATVGAAASLSDPNVLG--A
TH_3670|M.thermoresistible__bu IHVTLIEPGGYATDWGGSSARRSA-TMPEYAESRAEADRVRSQRVAR--P
MAV_2098|M.avium_104 VRVSVVEPAYTRTSFEAN-MADADAPIDAYAASREHARRRMAELMKT--A
MLBr_00862|M.leprae_Br4923 VGLTTICPGVIDTNIIQSTRIDTP----TAKQERVRDRRGQLDMMFRLRR
: : . *.
Mvan_0234|M.vanbaalenii_PYR-1 TDPAVVAELALDGLFAGQPEILADELTRNVKAQLSAAPA-----------
MSMEG_5711|M.smegmatis_MC2_155 SDPAMIATMAVDGLLGGDDEILADDWARTAKAQLSTRQ------------
Mb3199|M.bovis_AF2122/97 SDPADVVRQVLDGIEAGKEDVLADEMSRQVRASLNVPARERIARLMGN--
Rv3174|M.tuberculosis_H37Rv SDPADVVRQVLDGIEAGKEDVLADEMSRQVRASLNVPARERIARLMGN--
MAB_4428|M.abscessus_ATCC_1997 TPPSAIAARIIDALDKGDNEVLTDDLTVQIKSQLAGPVEKLAYRPGH---
MMAR_1614|M.marinum_M GDPAKAAAAIITAVESPAPPAFLLLGPDALMAYRHIAQRRDVEITD-WEQ
MUL_3423|M.ulcerans_Agy99 GDPAKAAAAIITAVESPAPPAFLLLGPDALMAYRHIAQRRDVEITD-WEQ
Mflv_0295|M.gilvum_PYR-GCK SDPAKVAGLLLDVIGMPDPPRRLLVGPDAYR-YATAAGRDLLAVDERWRT
TH_3670|M.thermoresistible__bu GDPKASAAALLKVVDAEKPPLRVFFG-ELPLQIVNADYQSRLQTWEEWQP
MAV_2098|M.avium_104 DDPAIVAEVVLKAATARKPKLRYPAGPLARRLSLLKKFAPEAVLDRGIRK
MLBr_00862|M.leprae_Br4923 YGPDKVADTILSAIKKNKPIRPVAPEAYALYGISRVLPQVLRSTARIRVI
* . :
Mvan_0234|M.vanbaalenii_PYR-1 -----------------------
MSMEG_5711|M.smegmatis_MC2_155 -----------------------
Mb3199|M.bovis_AF2122/97 -----------------------
Rv3174|M.tuberculosis_H37Rv -----------------------
MAB_4428|M.abscessus_ATCC_1997 -----------------------
MMAR_1614|M.marinum_M L---TAGTNLTP-----------
MUL_3423|M.ulcerans_Agy99 L---TAGTNLTP-----------
Mflv_0295|M.gilvum_PYR-GCK LSMSTAADDVSPEQLDPLGSASR
TH_3670|M.thermoresistible__bu IAVLAQG----------------
MAV_2098|M.avium_104 TNGLPAASRPHSHLQL-------
MLBr_00862|M.leprae_Br4923 -----------------------