For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
THDIVALVTGANRGLGRRLAAELVARGAKVYAAARRPETLDLPGVVPIPLDITDAESVRRAAEIAGDITV LVNNAGISTRANLLTGPVEDIRAEMETHYFGTLNVTRAFVPNIERNGGGAILNVLSVLSWMHPPTSGAYA SAKAAAWALTDAVRTELAPRGIHVAALHVGYMDTDMVPYVPADQKSDPAMIATMAVDGLLGGDDEILADD WARTAKAQLSTRQ*
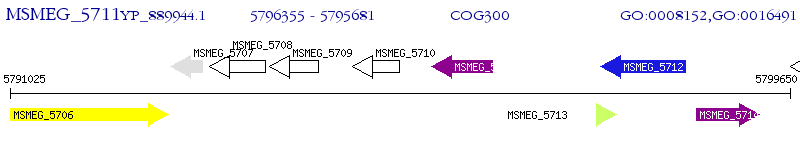
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5711 | - | - | 100% (224) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6581 | - | 2e-71 | 62.04% (216) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_1968 | - | 3e-56 | 53.92% (217) | putative oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3199 | - | 9e-48 | 45.87% (218) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0295 | - | 1e-23 | 38.12% (181) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv3174 | - | 3e-47 | 45.62% (217) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01094 | - | 6e-20 | 31.86% (204) | short chain alcohol dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4428 | - | 5e-43 | 43.24% (222) | short chain dehydrogenase |
| M. marinum M | MMAR_1614 | - | 7e-25 | 38.55% (179) | short chain dehydrogenase |
| M. avium 104 | MAV_2098 | - | 1e-23 | 34.81% (181) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_3670 | - | 4e-20 | 35.36% (181) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_3423 | - | 7e-25 | 38.55% (179) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0234 | - | 1e-95 | 77.27% (220) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_2098|M.avium_104 ---MTHRSPVVLVTGASSGIGRAVAKAFAAKGFEVFGTTRN---PRTAEP
Mflv_0295|M.gilvum_PYR-GCK ------MSKVLLISGASRGLGRAITESALAAGHLVVAGVRA---TSSLSD
TH_3670|M.thermoresistible__bu MGSNVWGSKVWFITGASRGFGREWAIAALDRGDRVAATARD---VSTLDD
MMAR_1614|M.marinum_M -------MSTWLITGCSTGLGRALAEAVIDAGHHTVATARS---VGGVAD
MUL_3423|M.ulcerans_Agy99 -------MSTWLITGCSTGLGRALAEAVIDAGHHTVATARS---VGGVAD
MSMEG_5711|M.smegmatis_MC2_155 ----MTHDIVALVTGANRGLGRRLAAELVARG-AKVYAAAR---RPETLD
Mvan_0234|M.vanbaalenii_PYR-1 --MTDTEHTTALVTGANRGLGRRFAEALVARG-AKVYAAAR---RPETID
Mb3199|M.bovis_AF2122/97 --MTSLAERTALVTGANRGMGREYVAQLLGRKVAKVYAATR---NPLAID
Rv3174|M.tuberculosis_H37Rv --MTSLAERTVLVTGANRGMGREYVAQLLGRKVAKVYAATR---NPLAID
MAB_4428|M.abscessus_ATCC_1997 ----MIKDTVVLVTGGRRGLGAAIVDEVLNRGARKVYSTAR---APFEDP
MLBr_01094|M.leprae_Br4923 --MEGFAGKVAVVTGAGSGIGQALAIELARSGAKLAISDVDGEGLAQTEG
. .::* *:* .
MAV_2098|M.avium_104 IPG-----VELVELDVTDAASVDAAAKSVIERTGRIDILVNNAGSGILGA
Mflv_0295|M.gilvum_PYR-GCK LATREPDRLAVVELDVTDDRHARAAVATAVERFGRLDVLVNNAGYANMSA
TH_3670|M.thermoresistible__bu LRDRFGDAVLAMTLDVTDRAADFAAVQRAHDHFGRLDIVVNNAGYGHFGF
MMAR_1614|M.marinum_M LVQRSPERVLPLALDITEPDQITAAVQAAQQRFGGIDVLVNNAGYGYRAA
MUL_3423|M.ulcerans_Agy99 LAQRSPERVLPLALDITEPDQITAAMQAAQQRFGGIDVLVNNAGYGYRAA
MSMEG_5711|M.smegmatis_MC2_155 L-----PGVVPIPLDITDAESVRRAAEIAGD----ITVLVNNAGISTRAN
Mvan_0234|M.vanbaalenii_PYR-1 I-----PGVVPIQLDITDPESVRRAAEQATD----VTVVINNAGVSTRAG
Mb3199|M.bovis_AF2122/97 VSD---PRVIPLQLDVTDAVSVAEAADLATD----VGILINNAGISRASS
Rv3174|M.tuberculosis_H37Rv VSD---PRVIPLQLDVTDAVSVAEAADLATD----VGILINNAGISRASS
MAB_4428|M.abscessus_ATCC_1997 R-----AQVVTKQLEVTSAESVDALARDLTD----VEIVVNNAGVLHPDP
MLBr_01094|M.leprae_Br4923 QLKAIGASARTDRLDVTEREAFLTYADVVHENFGKVNQIYNNAGIAFTGD
*::*. : : : ****
MAV_2098|M.avium_104 AEERSVAQAQRLIDTNFLGLVRLTNAVLPYLRAQGGGRVINIGSVLGFLP
Mflv_0295|M.gilvum_PYR-GCK IEDVDVDDFRAQIETNFFGVVNLTRAALPAMRRQRDGHIVQISSVGGRLV
TH_3670|M.thermoresistible__bu VEELTETEVRDQMETNFFGALWVTQAALPYLRRQRSGHIIQVSSIGGVSA
MMAR_1614|M.marinum_M VEEGDDAEVRDLFETHFFGTVALIKAVLPDMRARRSGAIVNISSIAVALT
MUL_3423|M.ulcerans_Agy99 VEEGDDAEVRDLFETHFFGTVALIKAVLPDMRARRSGAIVNISSIAVALT
MSMEG_5711|M.smegmatis_MC2_155 LLTGPVEDIRAEMETHYFGTLNVTRAFVPNIERNGGGAILNVLSVLSWMH
Mvan_0234|M.vanbaalenii_PYR-1 LLDGTLEDIRLEMDTHYFGTLAVTRAFVPVIERNGGGAILNVLSVLSWLH
Mb3199|M.bovis_AF2122/97 VLDKDTSALRGELETNLFGPLALASAFADRIAERSG-AIVNVSSVLAWLP
Rv3174|M.tuberculosis_H37Rv VLDKDTSALRGELETNLFGPLALASAFADRIAERSG-AIVNVSSVLAWLP
MAB_4428|M.abscessus_ATCC_1997 LLTGDFDRIDATFQTNVFGPLRIARAFAPILKANGGGAIVNIHSVLSWLG
MLBr_01094|M.leprae_Br4923 VEVSHFKDIERVMDVDYWGVVNGTKAFLPYLISSGDGHVINISSVFGLFS
::.. * : * : . :::: *:
MAV_2098|M.avium_104 QPYGALYAGSKHAVEGYSESLDHETREFG--VRVSVVEPAYTRTSFEAN-
Mflv_0295|M.gilvum_PYR-GCK RPGLAAYQSAKWAVTGFSGVLAVEVAPLG--IKVTVLEPGGMRTDWAGSS
TH_3670|M.thermoresistible__bu FADLGAYHASKWALEGLSQSLSLEVAPFG--IHVTLIEPGGYATDWGGSS
MMAR_1614|M.marinum_M PVGSGYYAAAKAAMEGMSGALHGELAPLG--ISVTVVEPGAFRTDFAGRS
MUL_3423|M.ulcerans_Agy99 PVGSGYYAAAKAAMEGMSGALHGELAPLG--ISVTVVEPGAFRTDFAGRS
MSMEG_5711|M.smegmatis_MC2_155 PPTSGAYASAKAAAWALTDAVRTELAPRG--IHVAALHVGYMDTDMVPY-
Mvan_0234|M.vanbaalenii_PYR-1 PPTSGAYSAAKAAGWAMTDALRAELAPRG--IHVAALHVGFMDTDMVSY-
Mb3199|M.bovis_AF2122/97 LGMS--YGVSKAAMWSATESMRIELAPRG--VQVVGVYVGLVDTDMGRF-
Rv3174|M.tuberculosis_H37Rv LGMS--YGVSKAAMWSATESMRIELAPRG--VQVVGVYVGLVDTDMGRF-
MAB_4428|M.abscessus_ATCC_1997 GAGA--YGASKAAIWSVSNTLRLELEAQH--THVLGVHAAFIDTDMVSA-
MLBr_01094|M.leprae_Br4923 VPGQAAYNSAKFAVRGFTEALREEMALAGRPVNVTTVYPGGIKTAIARNA
* :* * . : : * * : . *
MAV_2098|M.avium_104 MADADAPIDAYAASREHARR-RMAELMKTADDPAIVAEVVLKAATARKPK
Mflv_0295|M.gilvum_PYR-GCK MRIAP-VREEYAATVGAAASLSDPNVLG-ASDPAKVAGLLLDVIGMPDPP
TH_3670|M.thermoresistible__bu ARRSA-TMPEYAESRAEADRVRSQRVAR-PGDPKASAAALLKVVDAEKPP
MMAR_1614|M.marinum_M LVQSTRMINDYAATAGQRRKENDTMHGNQPGDPAKAAAAIITAVESPAPP
MUL_3423|M.ulcerans_Agy99 LVQSTRMINDYAATAGQRRKENDTMHGNQPGDPAKAAAAIITAVESPAPP
MSMEG_5711|M.smegmatis_MC2_155 ------------------------VPADQKSDPAMIATMAVDGLLGGDDE
Mvan_0234|M.vanbaalenii_PYR-1 ------------------------IPADQKTDPAVVAELALDGLFAGQPE
Mb3199|M.bovis_AF2122/97 ------------------------ADAP-KSDPADVVRQVLDGIEAGKED
Rv3174|M.tuberculosis_H37Rv ------------------------ADAP-KSDPADVVRQVLDGIEAGKED
MAB_4428|M.abscessus_ATCC_1997 ------------------------LPVP-KTPPSAIAARIIDALDKGDNE
MLBr_01094|M.leprae_Br4923 TAAEG---------LDVSKIASRFDTWVAHTSPQHAARIILKAVRKKKAR
* . :
MAV_2098|M.avium_104 LRYPAGPLARRLSLLKKFAPEAVLDRGIRKTNGLPAASRPHSHLQL----
Mflv_0295|M.gilvum_PYR-GCK RRLLVGPDAYRYATAAGRDLLAVDERWRTLSMSTAADDVSPEQLDPLGSA
TH_3670|M.thermoresistible__bu LRVFFGELPLQIVNADYQSRLQTWEEWQPIAVLAQG--------------
MMAR_1614|M.marinum_M AFLLLGPDALMAYRHIAQRRDVEITDWEQLTAGTNLTP------------
MUL_3423|M.ulcerans_Agy99 AFLLLGPDALMAYRHIAQRRDVEITDWEQLTAGTNLTP------------
MSMEG_5711|M.smegmatis_MC2_155 --ILADDWARTAKAQLSTRQ------------------------------
Mvan_0234|M.vanbaalenii_PYR-1 --ILADELTRNVKAQLSAAPA-----------------------------
Mb3199|M.bovis_AF2122/97 --VLADEMSRQVRASLNVPARERIARLMGN--------------------
Rv3174|M.tuberculosis_H37Rv --VLADEMSRQVRASLNVPARERIARLMGN--------------------
MAB_4428|M.abscessus_ATCC_1997 --VLTDDLTVQIKSQLAGPVEKLAYRPGH---------------------
MLBr_01094|M.leprae_Br4923 --VLVGPDAKVANVVVRFSGGAGYQRLFAQVASRLILNQR----------
. .
MAV_2098|M.avium_104 --
Mflv_0295|M.gilvum_PYR-GCK SR
TH_3670|M.thermoresistible__bu --
MMAR_1614|M.marinum_M --
MUL_3423|M.ulcerans_Agy99 --
MSMEG_5711|M.smegmatis_MC2_155 --
Mvan_0234|M.vanbaalenii_PYR-1 --
Mb3199|M.bovis_AF2122/97 --
Rv3174|M.tuberculosis_H37Rv --
MAB_4428|M.abscessus_ATCC_1997 --
MLBr_01094|M.leprae_Br4923 --