For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MIKDTVVLVTGGRRGLGAAIVDEVLNRGARKVYSTARAPFEDPRAQVVTKQLEVTSAESVDALARDLTDV EIVVNNAGVLHPDPLLTGDFDRIDATFQTNVFGPLRIARAFAPILKANGGGAIVNIHSVLSWLGGAGAYG ASKAAIWSVSNTLRLELEAQHTHVLGVHAAFIDTDMVSALPVPKTPPSAIAARIIDALDKGDNEVLTDDL TVQIKSQLAGPVEKLAYRPGH
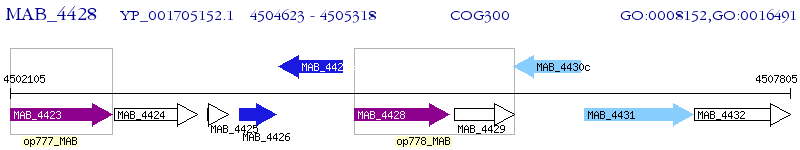
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4428 | - | - | 100% (231) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4335 | - | 2e-15 | 29.38% (211) | putative short-chain dehydrogenase/reductase |
| M. abscessus ATCC 19977 | MAB_1526 | - | 6e-15 | 30.77% (208) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3199 | - | 2e-40 | 38.30% (235) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4104 | - | 6e-16 | 30.84% (214) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv3174 | - | 7e-41 | 38.72% (235) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_02565 | fabG | 1e-16 | 30.57% (193) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. marinum M | MMAR_3561 | - | 5e-18 | 33.16% (193) | oxidoreductase |
| M. avium 104 | MAV_2098 | - | 2e-16 | 30.24% (248) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2899 | - | 3e-61 | 53.88% (219) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_4323 | - | 1e-15 | 31.41% (191) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_2794 | - | 4e-18 | 33.16% (193) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0234 | - | 7e-45 | 45.41% (218) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_4428|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_2899|M.smegmatis_MC2_155 --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
Mvan_0234|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_3561|M.marinum_M --------------------------------------------------
MUL_2794|M.ulcerans_Agy99 --------------------------------------------------
MAV_2098|M.avium_104 --------------------------------------------------
Mflv_4104|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_02565|M.leprae_Br4923 --------------------------------------------------
TH_4323|M.thermoresistible__bu MATEHFIDSSGGVRIAAYEEGNPDGPTVVLVHGWPDSHVVWDSVAPLLAD
MAB_4428|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_2899|M.smegmatis_MC2_155 --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
Mvan_0234|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_3561|M.marinum_M --------------------------------------------------
MUL_2794|M.ulcerans_Agy99 --------------------------------------------------
MAV_2098|M.avium_104 --------------------------------------------------
Mflv_4104|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_02565|M.leprae_Br4923 ----------------------------------------MAPKCSSDLF
TH_4323|M.thermoresistible__bu RFRIIRYDNRGVGRTTAPRRVADHTMARYADDFGAVIDALAPGQRVHVLA
MAB_4428|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_2899|M.smegmatis_MC2_155 --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
Mvan_0234|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_3561|M.marinum_M --------------------------------------------------
MUL_2794|M.ulcerans_Agy99 --------------------------------------------------
MAV_2098|M.avium_104 --------------------------------------------------
Mflv_4104|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_02565|M.leprae_Br4923 SQIVSSGPGSFVAKQLGVPRPETLRRYRPSDAPLCGSLLIGGDGRLVEPL
TH_4323|M.thermoresistible__bu HDWGSAGVWEYLARPEAEDRIASFTSVSGPSGDHLARYVLDGLAAPYRPR
MAB_4428|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_2899|M.smegmatis_MC2_155 --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
Mvan_0234|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_3561|M.marinum_M --------------------------------------------------
MUL_2794|M.ulcerans_Agy99 --------------------------------------------------
MAV_2098|M.avium_104 --------------------------------------------------
Mflv_4104|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_02565|M.leprae_Br4923 RATLEKDYDLVSNNLGGRWTDSFGGLVFDATGITAPAELKGLHNFFTPLL
TH_4323|M.thermoresistible__bu RFLRSLTQLLRLSYMGFFSIPVLAPLLVRGVLSKALRRSLRLRDGVPADR
MAB_4428|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_2899|M.smegmatis_MC2_155 --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
Mvan_0234|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_3561|M.marinum_M --------------------------------------------------
MUL_2794|M.ulcerans_Agy99 --------------------------------------------------
MAV_2098|M.avium_104 --------------------------------------------------
Mflv_4104|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_02565|M.leprae_Br4923 RNLGRCARVVVIGTTPDATTSTDERIAQRALEGFSRSLGKELRRGATVAL
TH_4323|M.thermoresistible__bu LHHSDTFAADAARSIKVYGANFLRTLRHARTDHFVRVPVQLIVNTRDPYV
MAB_4428|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_2899|M.smegmatis_MC2_155 --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
Mvan_0234|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_3561|M.marinum_M --------------------------------------------------
MUL_2794|M.ulcerans_Agy99 --------------------------------------------------
MAV_2098|M.avium_104 --------------------------------------------------
Mflv_4104|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_02565|M.leprae_Br4923 VYLSPDAKPAATGLESTMRFILSAKSAYVNGQVFYVGAADSTPPTN----
TH_4323|M.thermoresistible__bu RPHVYDETDRWVSRLWRRDIKAGHWSPMSHPQVLATAVTELADHLAGAPA
MAB_4428|M.abscessus_ATCC_1997 --------------MIKDTVVLVTGGRRGLGAAIVDEVLNRGA----RKV
MSMEG_2899|M.smegmatis_MC2_155 ------------MVHIAGRRMLVTGGQRGLGAAFASELLARGA----DRV
Mb3199|M.bovis_AF2122/97 ------------MTSLAERTALVTGANRGMGREYVAQLLGRKV----AKV
Rv3174|M.tuberculosis_H37Rv ------------MTSLAERTVLVTGANRGMGREYVAQLLGRKV----AKV
Mvan_0234|M.vanbaalenii_PYR-1 ------------MTDTEHTTALVTGANRGLGRRFAEALVARG-----AKV
MMAR_3561|M.marinum_M -------------MTSTPPVALITGVSSGIGAAIAVRLASAG-----FRV
MUL_2794|M.ulcerans_Agy99 -------------MTSTPPVALITGVSSGIGAAIAVRLASAG-----FRV
MAV_2098|M.avium_104 -------------MTHRSPVVLVTGASSGIGRAVAKAFAAKG-----FEV
Mflv_4104|M.gilvum_PYR-GCK --------------MAADRIALVTGAARGQGAAIVTRLLQDGFRVAACDL
MLBr_02565|M.leprae_Br4923 -----------WDRPLENKVAIVTGAARGIGAAIAEVVARDGAR-VVAID
TH_4323|M.thermoresistible__bu GRALLRAQVGRPREYFGDMLVAITGAGGGIGRETALAFAREGAEVIVSDI
:** * * . .
MAB_4428|M.abscessus_ATCC_1997 YSTARAP--FEDPRAQVVT--KQLEVTSAESVDALARDLTD-----VEIV
MSMEG_2899|M.smegmatis_MC2_155 YVTTRTP--APSSDARIVP--VALDVTDPDSVRAATEIADD-----VAIV
Mb3199|M.bovis_AF2122/97 YAATRNPLAIDVSDPRVIP--LQLDVTDAVSVAEAADLATD-----VGIL
Rv3174|M.tuberculosis_H37Rv YAATRNPLAIDVSDPRVIP--LQLDVTDAVSVAEAADLATD-----VGIL
Mvan_0234|M.vanbaalenii_PYR-1 YAAARRPETIDI--PGVVP--IQLDITDPESVRRAAEQATD-----VTVV
MMAR_3561|M.marinum_M VGTSRAP-QRLAPIPGVET--LALDVTDDTAVRSVVSEVIDRTG-RIDVL
MUL_2794|M.ulcerans_Agy99 VGTSRAP-QRLAPIPGVET--LALDVTDDTAVRSVVSEVIDRTG-HIDVL
MAV_2098|M.avium_104 FGTTRNP-RTAEPIPGVEL--VELDVTDAASVDAAAKSVIERTG-RIDIL
Mflv_4104|M.gilvum_PYR-GCK LGDDVTRTVAAHDTDHAVA--IELDVTSAEQWTRAVGEVVDRFG-GLSTL
MLBr_02565|M.leprae_Br4923 VESAAEALDETANRVGGTT--LSLDVTADDAVDKITEHLHDYQGGHADIL
TH_4323|M.thermoresistible__bu DEATAKDTAAEIAARGGVAHAYQLDVSDAAAVDRFADEVCARHG-VPDIV
*::: . :
MAB_4428|M.abscessus_ATCC_1997 VNNAGVLHPDPLLTGDFDRIDATFQTNVFGPLRIARAFAP-ILKANGGGA
MSMEG_2899|M.smegmatis_MC2_155 VNNAGTNGPLPVLGTDIDAMREVFETNVFGPVRVTQAFAP-ILARQPDSA
Mb3199|M.bovis_AF2122/97 INNAGISRASSVLDKDTSALRGELETNLFGPLALASAFAD-RIAERSG-A
Rv3174|M.tuberculosis_H37Rv INNAGISRASSVLDKDTSALRGELETNLFGPLALASAFAD-RIAERSG-A
Mvan_0234|M.vanbaalenii_PYR-1 INNAGVSTRAGLLDGTLEDIRLEMDTHYFGTLAVTRAFVP-VIERNGGGA
MMAR_3561|M.marinum_M VNNAGLGIAGAAEESSIDQARSLFDTNFFGLIRLTNEVLP-HMRRRGSGR
MUL_2794|M.ulcerans_Agy99 VNNAGLGIAGAAEESSIDQARSLFDTNFFGLIRLTNEVLP-HMRRRGSGR
MAV_2098|M.avium_104 VNNAGSGILGAAEERSVAQAQRLIDTNFLGLVRLTNAVLP-YLRAQGGGR
Mflv_4104|M.gilvum_PYR-GCK VNNAGVLHRASLADETPDGFEGSWRVNSLGPFLGIQAALP-HLKAAEGAA
MLBr_02565|M.leprae_Br4923 VNNAGITRDKLLANMDDCRWDAVLEVNLLAPQRLTEGLVG-NGSISKGGR
TH_4323|M.thermoresistible__bu VNNAGIGHAGRFLDTPRDQWDRVLEVNLGGVVNGCRAFAPRLVERGTGGH
:**** .: . .
MAB_4428|M.abscessus_ATCC_1997 IVNIHSVLSWLGGAGA--YGASKAAIWSVSNTLRLELEAQHTHVLGVHAA
MSMEG_2899|M.smegmatis_MC2_155 LVNVHSVFSWLGGTGA--YGASKAALWSLTNSLRVELAAQDTKVIGVHLS
Mb3199|M.bovis_AF2122/97 IVNVSSVLAWLPLGMS--YGVSKAAMWSATESMRIELAPRGVQVVGVYVG
Rv3174|M.tuberculosis_H37Rv IVNVSSVLAWLPLGMS--YGVSKAAMWSATESMRIELAPRGVQVVGVYVG
Mvan_0234|M.vanbaalenii_PYR-1 ILNVLSVLSWLHPPTSGAYSAAKAAGWAMTDALRAELAPRGIHVAALHVG
MMAR_3561|M.marinum_M IINISSVLGFLPAPYAALYAASKHAVEGYTESLDHELREYGVRALLVEPS
MUL_2794|M.ulcerans_Agy99 IINISSVLGFLPAPYAALYAASKHAVEGYTESLDHELREYGVRALLVEPS
MAV_2098|M.avium_104 VINIGSVLGFLPQPYGALYAGSKHAVEGYSESLDHETREFGVRVSVVEPA
Mflv_4104|M.gilvum_PYR-GCK IVNTCSTGAIRPFPNHAAYGSSKWALRGLTQVAAVELAPSGIRVNAVFPG
MLBr_02565|M.leprae_Br4923 VIVLSSMAGIAGNRGQTNYATAKAGVIGLTQALAPGLAEKGITINAVAPG
TH_4323|M.thermoresistible__bu IVNIASMASYAPLQSLNAYCTSKAAVYMFSDCLRAELDSAGIMLTTICPS
:: * . * :* . :: : .
MAB_4428|M.abscessus_ATCC_1997 FIDTDMVSALPVP-KTPPSAIAARIIDALDKGDNEVLTDDLTVQIKSQLA
MSMEG_2899|M.smegmatis_MC2_155 FADTDMTAGLDVF-KIAPEYAAARVLDGLERDDAEVLVDDTSRDVKAALA
Mb3199|M.bovis_AF2122/97 LVDTDMGRFADAP-KSDPADVVRQVLDGIEAGKEDVLADEMSRQVRASLN
Rv3174|M.tuberculosis_H37Rv LVDTDMGRFADAP-KSDPADVVRQVLDGIEAGKEDVLADEMSRQVRASLN
Mvan_0234|M.vanbaalenii_PYR-1 FMDTDMVSYIPADQKTDPAVVAELALDGLFAGQPEILADELTRNVKAQLS
MMAR_3561|M.marinum_M YTRTDFESNMWEADTPQAAYADNRAAVRAVVADKIRNAELPAVVADATYA
MUL_2794|M.ulcerans_Agy99 YTRTDFESNMWEADTPQAAYADNRAAVRAVVANKIRNAELPAVVADATYA
MAV_2098|M.avium_104 YTRTSFEANMADADAPIDAYAASREHARRRMAELMKTADDPAIVAEVVLK
Mflv_4104|M.gilvum_PYR-GCK PVETPMLDANTQTRLAANALMGRIGKPMEIADAVAFLVSEHATFITGSEL
MLBr_02565|M.leprae_Br4923 FIETQMTAAIPLATREVGRRLNSLLQGGLPVDVAETIAYFAIPASNAVTG
TH_4323|M.thermoresistible__bu TVNTNIIGSTRFNAPDGLDDVVDTRREQLQKLFALRRFGPDKAAKAIVSA
* :
MAB_4428|M.abscessus_ATCC_1997 GPVEKLAYRPGH--------------------------------------
MSMEG_2899|M.smegmatis_MC2_155 GPVGELVLTL----------------------------------------
Mb3199|M.bovis_AF2122/97 VPARERIARLMGN-------------------------------------
Rv3174|M.tuberculosis_H37Rv VPARERIARLMGN-------------------------------------
Mvan_0234|M.vanbaalenii_PYR-1 AAPA----------------------------------------------
MMAR_3561|M.marinum_M AATDKRIKLRYPAGRHARQLALLKRIAPAAVLDNGIRKQTGLNSP-RPTP
MUL_2794|M.ulcerans_Agy99 AAIDKRIKLRYPAGRHARQLALLKRIAPAAVLDNGIRKQTGLNSP-RPTP
MAV_2098|M.avium_104 AATARKPKLRYPAGPLARRLSLLKKFAPEAVLDRGIRKTNGLPAASRPHS
Mflv_4104|M.gilvum_PYR-GCK IVDGGQSLQIG---------------------------------------
MLBr_02565|M.leprae_Br4923 NVIRVCGQAMLGA-------------------------------------
TH_4323|M.thermoresistible__bu VKQNKPIRPVMPEAYFVYGVSRVLPQALRSTARGRVL-------------
MAB_4428|M.abscessus_ATCC_1997 -----
MSMEG_2899|M.smegmatis_MC2_155 -----
Mb3199|M.bovis_AF2122/97 -----
Rv3174|M.tuberculosis_H37Rv -----
Mvan_0234|M.vanbaalenii_PYR-1 -----
MMAR_3561|M.marinum_M VLAER
MUL_2794|M.ulcerans_Agy99 VLAER
MAV_2098|M.avium_104 HLQL-
Mflv_4104|M.gilvum_PYR-GCK -----
MLBr_02565|M.leprae_Br4923 -----
TH_4323|M.thermoresistible__bu -----