For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ELRNATVLVTGANRGLGAEFVRQVIGRGAAKVYAATRRPETITGGDSRIVPIYLDITDDATVQGAAGTAG DVDVVINNAGVMAMQNLVTGDLDAIRLEFATHFWGTLSMTRAFAPILARNGGGAILNVLSVGSFQVFPGN GAYAAAKAAEWQLANSTRLELAAQGTHVLGLHLAATDTDMLDGIDMPKNNPADVVRLALDGLESGLDEVL ADDETRTAKALLAQPPSAMYAGVEQ*
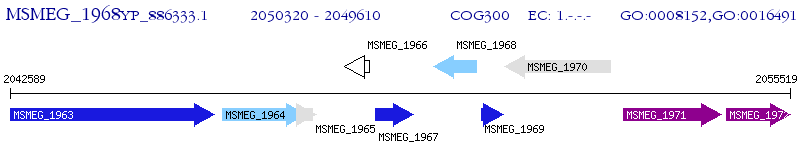
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1968 | - | - | 100% (236) | putative oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_5711 | - | 3e-56 | 53.92% (217) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_3805 | - | 2e-55 | 52.97% (236) | oxidoreductase, short chain dehydrogenase/reductase family |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3199 | - | 2e-48 | 45.54% (224) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0295 | - | 4e-20 | 33.33% (180) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv3174 | - | 5e-49 | 45.98% (224) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_00862 | ephD | 1e-12 | 28.87% (194) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4428 | - | 3e-48 | 45.98% (224) | short chain dehydrogenase |
| M. marinum M | MMAR_0810 | - | 2e-20 | 30.00% (230) | short-chain type oxidoreductase |
| M. avium 104 | MAV_4667 | - | 1e-18 | 34.25% (181) | oxidoreductase |
| M. thermoresistible (build 8) | TH_2589 | - | 6e-19 | 29.88% (241) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_4554 | - | 1e-20 | 30.00% (230) | short-chain type oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_0234 | - | 1e-57 | 54.39% (228) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1968|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0234|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
MAB_4428|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_0810|M.marinum_M --------------------------------------------------
MUL_4554|M.ulcerans_Agy99 --------------------------------------------------
MAV_4667|M.avium_104 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 MPATQSIHKVPHQFVESPDGVRLAIYQSGQPEGPAIVLVHGFPDSHVLWD
Mflv_0295|M.gilvum_PYR-GCK --------------------------------------------------
TH_2589|M.thermoresistible__bu --------------------------------------------------
MSMEG_1968|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0234|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
MAB_4428|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_0810|M.marinum_M --------------------------------------------------
MUL_4554|M.ulcerans_Agy99 --------------------------------------------------
MAV_4667|M.avium_104 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 GVVPLLAKRFRIVRYDNRGVGQSSVPKPVSAYTMTRFADDFAAVIDELSP
Mflv_0295|M.gilvum_PYR-GCK --------------------------------------------------
TH_2589|M.thermoresistible__bu --------------------------------------------------
MSMEG_1968|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0234|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
MAB_4428|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_0810|M.marinum_M --------------------------------------------------
MUL_4554|M.ulcerans_Agy99 --------------------------------------------------
MAV_4667|M.avium_104 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DRPVHVLAHDWGSVGVWEYLSRSGASDRVASFTSVSGPSQDQLVDYILSG
Mflv_0295|M.gilvum_PYR-GCK --------------------------------------------------
TH_2589|M.thermoresistible__bu --------------------------------------------------
MSMEG_1968|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0234|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
MAB_4428|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_0810|M.marinum_M --------------------------------------------------
MUL_4554|M.ulcerans_Agy99 --------------------------------------------------
MAV_4667|M.avium_104 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LRRPWRPRTFARAVSQLFRLTYMVLFSIPVLVPMVLWIALSSATIRRTMV
Mflv_0295|M.gilvum_PYR-GCK --------------------------------------------------
TH_2589|M.thermoresistible__bu --------------------------------------------------
MSMEG_1968|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0234|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
MAB_4428|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_0810|M.marinum_M --------------------------------------------------
MUL_4554|M.ulcerans_Agy99 --------------------------------------------------
MAV_4667|M.avium_104 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DNISTDQIHHSATLARDAAHSVKTYPANYFRAFSGRRRGKGVPVVNVPVQ
Mflv_0295|M.gilvum_PYR-GCK --------------------------------------------------
TH_2589|M.thermoresistible__bu --------------------------------------------------
MSMEG_1968|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0234|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
MAB_4428|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_0810|M.marinum_M --------------------------------------------------
MUL_4554|M.ulcerans_Agy99 --------------------------------------------------
MAV_4667|M.avium_104 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LIVNTMDRYVRPYGYDATARWVPRLWRRDIKAGHFSPMSHPQVMAAAVHD
Mflv_0295|M.gilvum_PYR-GCK --------------------------------------------------
TH_2589|M.thermoresistible__bu --------------------------------------------------
MSMEG_1968|M.smegmatis_MC2_155 -----------------------MELRNATVLVTGANRGLGAEFVRQVIG
Mvan_0234|M.vanbaalenii_PYR-1 ----------------------MTDTEHTTALVTGANRGLGRRFAEALVA
Mb3199|M.bovis_AF2122/97 ----------------------MTSLAERTALVTGANRGMGREYVAQLLG
Rv3174|M.tuberculosis_H37Rv ----------------------MTSLAERTVLVTGANRGMGREYVAQLLG
MAB_4428|M.abscessus_ATCC_1997 ------------------------MIKDTVVLVTGGRRGLGAAIVDEVLN
MMAR_0810|M.marinum_M --------------------MTTTDAGNRVAVVTGASSGIGEATARTLAA
MUL_4554|M.ulcerans_Agy99 --------------------MTTTDAGNRVAVVTGASSGIGEATARTLAA
MAV_4667|M.avium_104 --------------------MVGTGRNERVAVVTGASSGIGEATARTLAA
MLBr_00862|M.leprae_Br4923 LADLVDGKQPSRALLRAQVGRSRDTFGDTLVSVTGAGSGIGRETALALAR
Mflv_0295|M.gilvum_PYR-GCK --------------------------MSKVLLISGASRGLGRAITESALA
TH_2589|M.thermoresistible__bu --------------VDRQTFDKLFDMTGRTVVVTGGTRGIGLALAEGYVL
::*. *:* .
MSMEG_1968|M.smegmatis_MC2_155 RGAAKVYA-----ATRRPETITGGDSR-IVPIYLDITDDATVQGAAGTAG
Mvan_0234|M.vanbaalenii_PYR-1 RG-AKVYA-----AARRPETIDIPG---VVPIQLDITDPESVRRAAEQAT
Mb3199|M.bovis_AF2122/97 RKVAKVYA-----ATRNPLAIDVSDPR-VIPLQLDVTDAVSVAEAADLAT
Rv3174|M.tuberculosis_H37Rv RKVAKVYA-----ATRNPLAIDVSDPR-VIPLQLDVTDAVSVAEAADLAT
MAB_4428|M.abscessus_ATCC_1997 RGARKVYS-----TARAP--FEDPRAQ-VVTKQLEVTSAESVDALARDLT
MMAR_0810|M.marinum_M QGFHVVAV-----ARRADRITALTDQIGGTPIVADVTDGAAVAALADGLS
MUL_4554|M.ulcerans_Agy99 QGFHVVAV-----ARRADRITALTDQIGGTPIVADVTDGAAVAALADGLS
MAV_4667|M.avium_104 QGFHVVAV-----ARRAKRIHALANEIGGTAIVADVTDDAAVAALASKLS
MLBr_00862|M.leprae_Br4923 RGAEVVVSDIDEAAAKNTAAQIVASGGVAYAYALDVSDAAAVEAFAEQVS
Mflv_0295|M.gilvum_PYR-GCK AGHLVVAG---VRATSSLSDLATREPDRLAVVELDVTDDRHARAAVATAV
TH_2589|M.thermoresistible__bu AGARVVVASRKSDACERAAAHLRALGGDAIGVPTHLGDIDACEALVRRTV
* : .: . .
MSMEG_1968|M.smegmatis_MC2_155 DVD----VVINNAG-VMAMQNLVTGDLDAIRLEFATHFWGTLSMTRAFAP
Mvan_0234|M.vanbaalenii_PYR-1 DVT----VVINNAG-VSTRAGLLDGTLEDIRLEMDTHYFGTLAVTRAFVP
Mb3199|M.bovis_AF2122/97 DVG----ILINNAG-ISRASSVLDKDTSALRGELETNLFGPLALASAFAD
Rv3174|M.tuberculosis_H37Rv DVG----ILINNAG-ISRASSVLDKDTSALRGELETNLFGPLALASAFAD
MAB_4428|M.abscessus_ATCC_1997 DVE----IVVNNAG-VLHPDPLLTGDFDRIDATFQTNVFGPLRIARAFAP
MMAR_0810|M.marinum_M RVD----VLVNNAGGAKGLEPVADADLEHWRWMWETNVLGTLRVTRALLP
MUL_4554|M.ulcerans_Agy99 RVD----VLVNNAGGAKELEPVADADLEHWRWMWETNVLGTLRVTRALLP
MAV_4667|M.avium_104 RVD----VLVNNAGGARGLEPVAEADLEHWRWMWETNVIGTLRVTRALLP
MLBr_00862|M.leprae_Br4923 AKHGVPDIVVNNAG-IGQAGRFLDTPPEEFDRVLAVNLGGVVNCCRAFGQ
Mflv_0295|M.gilvum_PYR-GCK ERFGRLDVLVNNAG-YANMSAIEDVDVDDFRAQIETNFFGVVNLTRAALP
TH_2589|M.thermoresistible__bu DEFGGIDVLVNNAA-NALTQPLGELTVDAWTKSFEVNLRGPVFLVQAALP
:::***. . . .: * : *
MSMEG_1968|M.smegmatis_MC2_155 ILAR-NGGGAILNVLSVGSFQVFPGNGAYAAAKAAEWQLANSTRLELAAQ
Mvan_0234|M.vanbaalenii_PYR-1 VIER-NGGGAILNVLSVLSWLHPPTSGAYSAAKAAGWAMTDALRAELAPR
Mb3199|M.bovis_AF2122/97 RIAE-RSG-AIVNVSSVLAWLPLGMS--YGVSKAAMWSATESMRIELAPR
Rv3174|M.tuberculosis_H37Rv RIAE-RSG-AIVNVSSVLAWLPLGMS--YGVSKAAMWSATESMRIELAPR
MAB_4428|M.abscessus_ATCC_1997 ILKA-NGGGAIVNIHSVLSWL--GGAGAYGASKAAIWSVSNTLRLELEAQ
MMAR_0810|M.marinum_M KLIA-SGDGLIVTVTSIAALEIYDGGAGYTAAKHAQGALHRTLRGELLGL
MUL_4554|M.ulcerans_Agy99 KLIA-SGDGLIVTVTSIAALEIYDGGAGYTAAKHAQGALHRTLRGELLGL
MAV_4667|M.avium_104 KLVD-SGDGLIVTVTSVAALEVYDGGAGYTAAKHGQGALHRTLRGELLGK
MLBr_00862|M.leprae_Br4923 RLVERGTGGHIVNVSSMAAYAPLQSLSAYCTSKAATYMFSDCLRAELDAV
Mflv_0295|M.gilvum_PYR-GCK AMRR-QRDGHIVQISSVGGRLVRPGLAAYQSAKWAVTGFSGVLAVEVAPL
TH_2589|M.thermoresistible__bu HLKT-SPHAAVLNMVSVGAFLFSPGTAMYSAGKAALMSFTRSMAAQFAPL
: :: : *: . * .* . :.
MSMEG_1968|M.smegmatis_MC2_155 GTHVLGLHLAATDTDMLDGIDMP-KNNPADVVR-------LALDGLESGL
Mvan_0234|M.vanbaalenii_PYR-1 GIHVAALHVGFMDTDMVSYIPADQKTDPAVVAE-------LALDGLFAGQ
Mb3199|M.bovis_AF2122/97 GVQVVGVYVGLVDTDMGRFADAP-KSDPADVVR-------QVLDGIEAGK
Rv3174|M.tuberculosis_H37Rv GVQVVGVYVGLVDTDMGRFADAP-KSDPADVVR-------QVLDGIEAGK
MAB_4428|M.abscessus_ATCC_1997 HTHVLGVHAAFIDTDMVSALPVP-KTPPSAIAA-------RIIDALDKGD
MMAR_0810|M.marinum_M PVRLTEIAPGAVETEFSLVRFDGDEQRADAVYS-------GMTPLVAADI
MUL_4554|M.ulcerans_Agy99 PVRLTEIAPGAVETEFSLVRFDGDEQRADAVYS-------GMTPLVAADI
MAV_4667|M.avium_104 PVRLTEIAPGAVETEFSLVRFDGDQQRADAVYT-------GITPLVAADV
MLBr_00862|M.leprae_Br4923 GVGLTTICPGVIDTNIIQSTRIDTPTAKQERVRDRRGQLDMMFRLRRYGP
Mflv_0295|M.gilvum_PYR-GCK GIKVTVLEPGGMRTDWAGSSMRIAPVREEYAATVGAAASLSDPNVLGASD
TH_2589|M.thermoresistible__bu GIRVNALAPGPVDTDMMRNNPQE---VVDMMAG-------ATLMKRLATP
: : . *:
MSMEG_1968|M.smegmatis_MC2_155 DEVLADDETRTA------KALLAQPPSAMYAGVEQ---------------
Mvan_0234|M.vanbaalenii_PYR-1 PEILADELTRNV------KAQLSAAPA-----------------------
Mb3199|M.bovis_AF2122/97 EDVLADEMSRQV------RASLNVPARERIARLMGN--------------
Rv3174|M.tuberculosis_H37Rv EDVLADEMSRQV------RASLNVPARERIARLMGN--------------
MAB_4428|M.abscessus_ATCC_1997 NEVLTDDLTVQI------KSQLAGPVEKLAYRPGH---------------
MMAR_0810|M.marinum_M AEVIGFVASRPP------HVNLDQIIIRPRDQASATRRANHQV-------
MUL_4554|M.ulcerans_Agy99 AEVIGFVASRPP------HVNLDQIIIRPRDQASATRRANHQV-------
MAV_4667|M.avium_104 AEVIGFVASRPA------HVNLDLIVMKPRDQASASRFNRRG--------
MLBr_00862|M.leprae_Br4923 DKVADTILSAIK------KNKPIRPVAPEAYALYGISRVLPQVLRSTARI
Mflv_0295|M.gilvum_PYR-GCK PAKVAGLLLDVIGMPDPPRRLLVGPDAYRYATAAGRDLLAVDERWRTLSM
TH_2589|M.thermoresistible__bu DEMVGAALLLTS------DAGSYITGTVLIADGGGTPR------------
MSMEG_1968|M.smegmatis_MC2_155 --------------------
Mvan_0234|M.vanbaalenii_PYR-1 --------------------
Mb3199|M.bovis_AF2122/97 --------------------
Rv3174|M.tuberculosis_H37Rv --------------------
MAB_4428|M.abscessus_ATCC_1997 --------------------
MMAR_0810|M.marinum_M --------------------
MUL_4554|M.ulcerans_Agy99 --------------------
MAV_4667|M.avium_104 --------------------
MLBr_00862|M.leprae_Br4923 RVI-----------------
Mflv_0295|M.gilvum_PYR-GCK STAADDVSPEQLDPLGSASR
TH_2589|M.thermoresistible__bu --------------------