For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
STWLITGCSTGLGRALAEAVIDAGHHTVATARSVGGVADLVQRSPERVLPLALDITEPDQITAAVQAAQQ RFGGIDVLVNNAGYGYRAAVEEGDDAEVRDLFETHFFGTVALIKAVLPDMRARRSGAIVNISSIAVALTP VGSGYYAAAKAAMEGMSGALHGELAPLGISVTVVEPGAFRTDFAGRSLVQSTRMINDYAATAGQRRKEND TMHGNQPGDPAKAAAAIITAVESPAPPAFLLLGPDALMAYRHIAQRRDVEITDWEQLTAGTNLTP*
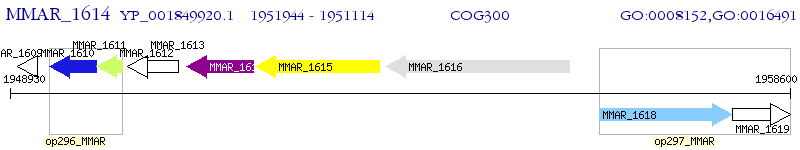
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1614 | - | - | 100% (276) | short chain dehydrogenase |
| M. marinum M | MMAR_3561 | - | 3e-31 | 41.94% (186) | oxidoreductase |
| M. marinum M | MMAR_4002 | - | 3e-25 | 32.16% (255) | dehydrogenase/decarboxylase protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1385 | fabG | 7e-22 | 33.52% (182) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. gilvum PYR-GCK | Mflv_0295 | - | 6e-52 | 44.28% (271) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv1350 | fabG | 7e-22 | 33.52% (182) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. leprae Br4923 | MLBr_01094 | - | 2e-17 | 26.69% (251) | short chain alcohol dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1305 | - | 4e-26 | 32.79% (247) | short-chain dehydrogenase/reductase |
| M. avium 104 | MAV_3827 | - | 2e-67 | 49.82% (273) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2026 | - | 1e-111 | 72.16% (273) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_3670 | - | 1e-46 | 40.30% (268) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_3423 | - | 1e-154 | 99.28% (276) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_3508 | - | 5e-51 | 40.00% (275) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1385|M.bovis_AF2122/97 -MASLLNARTAVITGGAQG-LGLAIGQRFVAEGARVVLGDVNLEATEVAA
Rv1350|M.tuberculosis_H37Rv -MASLLNARTAVITGGAQG-LGLAIGQRFVAEGARVVLGDVNLEATEVAA
MMAR_1614|M.marinum_M -------MSTWLITGCSTG-LGRALAEAVIDAGHHTVATARSVGGVADLV
MUL_3423|M.ulcerans_Agy99 -------MSTWLITGCSTG-LGRALAEAVIDAGHHTVATARSVGGVADLA
MSMEG_2026|M.smegmatis_MC2_155 -------MTTWLITGCSTG-IGRTLAEAVAAAGHNLVVTARDVSSVADIA
MAV_3827|M.avium_104 -------MARWLITGCSTG-LGREIAGAALQAGHRVVVTARRADAVRGFA
Mvan_3508|M.vanbaalenii_PYR-1 -------MRTWFITGGTPGGFGMAFADAALDAGDRVALTVRRPEELDGWA
Mflv_0295|M.gilvum_PYR-GCK ------MSKVLLISGASRG-LGRAITESALAAGHLVVAGVRATSSLSDLA
TH_3670|M.thermoresistible__bu MGSNVWGSKVWFITGASRG-FGREWAIAALDRGDRVAATARDVSTLDDLR
MAB_1305|M.abscessus_ATCC_1997 ----MSEHWTVLVTGASSG-IGASAARIFAERGHRVFGTSRNPDTITDRV
MLBr_01094|M.leprae_Br4923 --MEGFAGKVAVVTGAGSG-IGQALAIELARSGAKLAISDVDGEGLAQTE
.::* * :* *
Mb1385|M.bovis_AF2122/97 KRLGGD-DVALAVRCDVTQADDVDILIRTAVERFGGLDVMVNNAGITRDA
Rv1350|M.tuberculosis_H37Rv KRLGGD-DVALAVRCDVTQADDVDILIRTAVERFGGLDVMVNNAGITRDA
MMAR_1614|M.marinum_M QRSP---ERVLPLALDITEPDQITAAVQAAQQRFGGIDVLVNNAGYGYRA
MUL_3423|M.ulcerans_Agy99 QRSP---ERVLPLALDITEPDQITAAMQAAQQRFGGIDVLVNNAGYGYRA
MSMEG_2026|M.smegmatis_MC2_155 ETAP---DRVVAAPLDVTRPEQVTAAAQLADERFGGVDVLVNNAGYGYRA
MAV_3827|M.avium_104 EEFG---ELALPVALDVTDRDQIAAAVAAADAAFGGIDVLVNNAGHGYLS
Mvan_3508|M.vanbaalenii_PYR-1 REHG---DRVLVVPLDVTDPEQVRVAVKATEERFGGIDVLVNNAGRAWAG
Mflv_0295|M.gilvum_PYR-GCK TREP---DRLAVVELDVTDDRHARAAVATAVERFGRLDVLVNNAGYANMS
TH_3670|M.thermoresistible__bu DRFG---DAVLAMTLDVTDRAADFAAVQRAHDHFGRLDIVVNNAGYGHFG
MAB_1305|M.abscessus_ATCC_1997 PGVQ-------YLRLDQTDK----ASIAECVAQAGEVDILVNNAGESQIG
MLBr_01094|M.leprae_Br4923 GQLKAIGASARTDRLDVTEREAFLTYADVVHENFGKVNQIYNNAGIAFTG
* * * :: : **** .
Mb1385|M.bovis_AF2122/97 TMRTMTEEQFDQVIAVHLKGTWNGTRLAAAIMRERKRGAIVNMSSVSGKV
Rv1350|M.tuberculosis_H37Rv TMRTMTEEQFDQVIAVHLKGTWNGTRLAAAIMRERKRGAIVNMSSVSGKV
MMAR_1614|M.marinum_M AVEEGDDAEVRDLFETHFFGTVALIKAVLPDMRARRSGAIVNISSIAVAL
MUL_3423|M.ulcerans_Agy99 AVEEGDDAEVRDLFETHFFGTVALIKAVLPDMRARRSGAIVNISSIAVAL
MSMEG_2026|M.smegmatis_MC2_155 AVEEGDDADVRTLFDTHIFGTVATIKAVLPGMRARRRGAIVNISSIGAQI
MAV_3827|M.avium_104 AVEEGEDAEVRKLFDVNYFGAVDMIKAVLPGMRARGCGHIVNISSMTGLV
Mvan_3508|M.vanbaalenii_PYR-1 TVEGMDESELRAMFELNFFAVLSVLRAALPGMRARRGGWIVNVSSVAGLR
Mflv_0295|M.gilvum_PYR-GCK AIEDVDVDDFRAQIETNFFGVVNLTRAALPAMRRQRDGHIVQISSVGGRL
TH_3670|M.thermoresistible__bu FVEELTETEVRDQMETNFFGALWVTQAALPYLRRQRSGHIIQVSSIGGVS
MAB_1305|M.abscessus_ATCC_1997 ALEDVSMDDFEKLYMTNVFGPVALTKAVLPGMRERRRGAVVMVGSMAGTL
MLBr_01094|M.leprae_Br4923 DVEVSHFKDIERVMDVDYWGVVNGTKAFLPYLISSGDGHVINISSVFGLF
:. :. . . : . : * :: :.*:
Mb1385|M.bovis_AF2122/97 GMVGQTNYSAAKAGIVGMTKAAAKELAHLG--IRVNAIAPGLIRS-----
Rv1350|M.tuberculosis_H37Rv GMVGQTNYSAAKAGIVGMTKAAAKELAHLG--IRVNAIAPGLIRS-----
MMAR_1614|M.marinum_M TPVGSGYYAAAKAAMEGMSGALHGELAPLG--ISVTVVEPGAFRT-DFAG
MUL_3423|M.ulcerans_Agy99 TPVGSGYYAAAKAAMEGMSGALHGELAPLG--ISVTVVEPGAFRT-DFAG
MSMEG_2026|M.smegmatis_MC2_155 TPPGSGYYAAAKAAVEGLSGSLHTELAPLG--ISVTVVEPGGFRT-DFAG
MAV_3827|M.avium_104 ANPPNAYYSSTKFALEAVTEALAAEVRPLG--IKVTAIEPGAFRT-DWAT
Mvan_3508|M.vanbaalenii_PYR-1 GVTGFGHYSATKFAVEAITGVLRDEVAPWG--INVMAVEPAAFRTRAYSG
Mflv_0295|M.gilvum_PYR-GCK VRPGLAAYQSAKWAVTGFSGVLAVEVAPLG--IKVTVLEPGGMRT-DWAG
TH_3670|M.thermoresistible__bu AFADLGAYHASKWALEGLSQSLSLEVAPFG--IHVTLIEPGGYAT-DWGG
MAB_1305|M.abscessus_ATCC_1997 HVPFRSTYSSAKSALHTVADCLRYEVAPFG--ITVSTVEPGVIAT---GI
MLBr_01094|M.leprae_Br4923 SVPGQAAYNSAKFAVRGFTEALREEMALAGRPVNVTTVYPGGIKT-----
* ::* .: .: *: * : * : *. :
Mb1385|M.bovis_AF2122/97 --------AMTEAMPQRIWDQKLAEVPMG-RAGEPSEVASVAVFLASDLS
Rv1350|M.tuberculosis_H37Rv --------AMTEAMPQRIWDQKLAEVPMG-RAGEPSEVASVAVFLASDLS
MMAR_1614|M.marinum_M RSLVQSTRMINDYAATAGQRRKENDTMHGNQPGDPAKAAAAIITAVESPA
MUL_3423|M.ulcerans_Agy99 RSLVQSTRMINDYAATAGQRRKENDTMHGNQPGDPAKAAAAIITAVESPA
MSMEG_2026|M.smegmatis_MC2_155 RSLKQSAAAIDDYADTAGKRRKEHDRVHGTQPGDPVKAAAAIIKAVESPE
MAV_3827|M.avium_104 RSMKESGTPIADYTDVAA-RKDLIKQFADHLPGDPRKVAEAVLMVTGLDE
Mvan_3508|M.vanbaalenii_PYR-1 FADEPLREPVSEYRALLQGVREAMIAQDGMQPGDPRRGARAVLAAMAQDP
Mflv_0295|M.gilvum_PYR-GCK SSMR-IAPVREEYAATVGAAASLS-DPNVLGASDPAKVAGLLLDVIGMPD
TH_3670|M.thermoresistible__bu SSAR-RSATMPEYAESRAEADRVR-SQRVARPGDPKASAAALLKVVDAEK
MAB_1305|M.abscessus_ATCC_1997 ETRRNRIVPDGSPYRDAFRTVDAAMAAREEHGTATDELGRLVVDVALSPS
MLBr_01094|M.leprae_Br4923 -----AIARNATAAEGLDVSKIASRFDTWVAHTSPQHAARIILKAVR--K
. . :
Mb1385|M.bovis_AF2122/97 S----YMTGTVLDVTGGRFI------------------------------
Rv1350|M.tuberculosis_H37Rv S----YMTGTVLDVTGGRFI------------------------------
MMAR_1614|M.marinum_M PPAFLLLGPDALMAYRHIAQRRDVEITDWEQLTAGTNLTP----------
MUL_3423|M.ulcerans_Agy99 PPAFLLLGPDALMAYRHIAQRRDVEITDWEQLTAGTNLTP----------
MSMEG_2026|M.smegmatis_MC2_155 PPAFLLLGSDALSAYRRVAQARAEEIDKWADLTTSTDFDS----------
MAV_3827|M.avium_104 PPLRLLLGRDVLKAMRDKIAAMSASIDEWEAVTKDVNFPGA---------
Mvan_3508|M.vanbaalenii_PYR-1 PPHRLVLGGDGFDAVKAELEAALAELHENETLSRGADFPH----------
Mflv_0295|M.gilvum_PYR-GCK PPRRLLVGPDAYRYATAAGRDLLAVDERWRTLSMSTAADDVSPEQLDPLG
TH_3670|M.thermoresistible__bu PPLRVFFGELPLQIVNADYQSRLQTWEEWQPIAVLAQG------------
MAB_1305|M.abscessus_ATCC_1997 PKPLYAKGSNAPIIMTAQRLLPARLFLKFLARMHGLKVR-----------
MLBr_01094|M.leprae_Br4923 KKARVLVGPDAKVANVVVRFSGGAGYQRLFAQVASRLILNQR--------
Mb1385|M.bovis_AF2122/97 ----
Rv1350|M.tuberculosis_H37Rv ----
MMAR_1614|M.marinum_M ----
MUL_3423|M.ulcerans_Agy99 ----
MSMEG_2026|M.smegmatis_MC2_155 ----
MAV_3827|M.avium_104 ----
Mvan_3508|M.vanbaalenii_PYR-1 ----
Mflv_0295|M.gilvum_PYR-GCK SASR
TH_3670|M.thermoresistible__bu ----
MAB_1305|M.abscessus_ATCC_1997 ----
MLBr_01094|M.leprae_Br4923 ----