For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TSMVQTTDLTGQMLIAGTPVRGAGREIRGIDPQTGSFLEPAYPYGDTSHVEAACAAAADAFGPYRNAPVE QRARFLEAIADNLDSSRDVLVERAHAESGLPVARLTGEVGRTSGQLRLFAGVLREGSWNQARIDPALPDR EPLPRPDIRQRSVPLGPVAVFGASNFPLAFSVAGGDTASALAAGCPVVVKAHDAHPGTSELVGRAIAQAV TATGMPAGTFSLLYGYGPDLGSALVTDPRIKAVGFTGSRAGGMALVAAAAARPEPIPVYAEMSAINPVFL LRGALSARAADLARAFVGSLTMGSGQFCTNPGLVIAVDGPDLDAFVAAAAEAVAATAPTPMLTPGIAENF RTGVETLSGTAELIARGSDENAPAVSCRAALFGSDVPTFLATEALQAEVFGASGVIVRCADDAEMLRVAA EVEGQLTATVHAEESDHAKAGELLEILELKAGRILFNGWPTGVEVGHAMVHGGPFPSTSDSRTTSVGARA IERFLRPVCYQNAPKSLLPSAIADGNPDGMWRTVDGQLTKD*
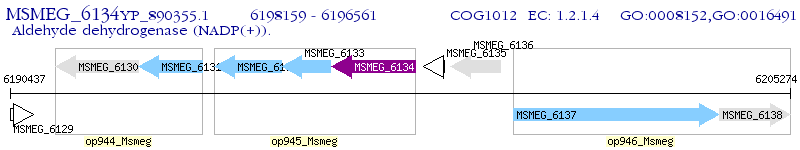
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6134 | - | - | 100% (532) | NADP-dependent fatty aldehyde dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6874 | - | 5e-26 | 27.83% (442) | aldehyde dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6687 | - | 7e-26 | 29.41% (442) | aldehyde dehydrogenase, thermostable |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0228c | - | 2e-20 | 29.29% (338) | aldehyde dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0428 | - | 3e-21 | 26.22% (450) | betaine-aldehyde dehydrogenase |
| M. tuberculosis H37Rv | Rv0223c | - | 1e-21 | 28.29% (410) | aldehyde dehydrogenase |
| M. leprae Br4923 | MLBr_02573 | gabD1 | 2e-09 | 27.59% (290) | succinic semialdehyde dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3328 | - | 2e-23 | 28.21% (475) | gamma-aminobutyraldehyde dehydrogenase |
| M. marinum M | MMAR_2610 | - | 1e-23 | 27.61% (460) | aldehyde dehydrogenase |
| M. avium 104 | MAV_2101 | - | 5e-21 | 25.64% (433) | aldehyde dehydrogenase |
| M. thermoresistible (build 8) | TH_4531 | - | 9e-30 | 29.53% (491) | PUTATIVE putative betaine aldehyde dehydrogenase |
| M. ulcerans Agy99 | MUL_2789 | - | 1e-20 | 26.23% (488) | gamma-aminobutyraldehyde dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_1031 | - | 0.0 | 71.67% (533) | aldehyde dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_3328|M.abscessus_ATCC_1997 --------MSVPSSWINGRPVATH-GDTHRVINPATGEAVADLKLA-TTA
MUL_2789|M.ulcerans_Agy99 ----MTTTVSVAGSWINGAAHSTG-GRQHAVINPATQETIAELALA-TPA
MMAR_2610|M.marinum_M -----MVEKKVFHNIIGGEIQATPDGNTMDIVNPSTGEVYASAPNS-SGK
MLBr_02573|M.leprae_Br4923 --------------------------MSIATINPATGETVKTFTPT-TQD
Mb0228c|M.bovis_AF2122/97 MSDS---ATEYDKLFIGGKWTKPSTSDVIEVRCPATGEYVGKVPMA-AAA
Rv0223c|M.tuberculosis_H37Rv MSDS---ATEYDKLFIGGKWTKPSTSDVIEVRCPATGEYVGKVPMA-AAA
Mflv_0428|M.gilvum_PYR-GCK MTQSTAFRTEWDKLFIGGKWVEPASSEVIEVRSPATGDVVGKVPLA-SAA
MAV_2101|M.avium_104 ------------------------MSTTIPVFNPSTEEQIAEVPDS-DQA
TH_4531|M.thermoresistible__bu --VTTTENRLVSEIHLAGRRLAEGSAGVYEHVNPATGKVQGEVPLA-GPA
MSMEG_6134|M.smegmatis_MC2_155 MTSMVQ-TTDLTGQMLIAGTPVRGAGREIRGIDPQTGSFLEPAYPYGDTS
Mvan_1031|M.vanbaalenii_PYR-1 MTATISGATSLTGQMLIAGAPVRGTGKEVRAFDPAAGQPLEPVYQHGDNT
* : .
MAB_3328|M.abscessus_ATCC_1997 DVDAAVAAARAAGPA--WASSTPVDRSSVLAKLAAVLSANADTLVADEVA
MUL_2789|M.ulcerans_Agy99 DVDHAVVAARRALPQ--WAGVTTAERSAVLAELADLLEQRAPELIAEEVC
MMAR_2610|M.marinum_M DVDDAFTAAAKAFES--WRWSTPSERQRALLRLADLIEDNAEELVALECE
MLBr_02573|M.leprae_Br4923 EVEDAIARAYARFDN--YRHTTFTQRAKWANATADLLEAEANQSAALMTL
Mb0228c|M.bovis_AF2122/97 DVDAAVAAARAAFDNGPWPSTPPHERAAVIAAAVKMLAERKDLFTKLLAA
Rv0223c|M.tuberculosis_H37Rv DVDAAVAAARAAFDNGPWPSTPPHERAAVIAAAVKMLAERKDLFTKLLAA
Mflv_0428|M.gilvum_PYR-GCK DVDAACAAAREAFDNGPWPQMSPTERAEVLGRAVKLMEERADELKFLLAA
MAV_2101|M.avium_104 AVDAAVARARETFESGVWRKAPASHRANVLFRAARIIEERKDELAALESQ
TH_4531|M.thermoresistible__bu EVDEAVAAAQEAFKE--WRKWRPEDRRRLLTRIAERIRAHEDELAVLQAR
MSMEG_6134|M.smegmatis_MC2_155 HVEAACAAAADAFGP--YRNAPVEQRARFLEAIADNLDSSRDVLVERAHA
Mvan_1031|M.vanbaalenii_PYR-1 HVDAACAAAADAFAQ--YRATTSEQRAAFLDTIATNIEAVGEALIARAVA
*: * . * : .* . :
MAB_3328|M.abscessus_ATCC_1997 QTGKPVRLASEFDVPGSIDNVEFFAGAARHLEGKAT-----AEYSAD--H
MUL_2789|M.ulcerans_Agy99 QTGKPLRLAEEFDVPGSVDNIRFFAGAARRLPGQAT-----AEYSAD--H
MMAR_2610|M.marinum_M NTGKPISLTMSEEIPPMVDQIRFFAGAARVLEGRSQ-----GEYMRG--H
MLBr_02573|M.leprae_Br4923 EMGKTLQSAKDEAMK-CAKGFRYYAEKAETLLADEP-----AEAAAVGAS
Mb0228c|M.bovis_AF2122/97 ETGQPPTIIETMHWMGSMGAMNYFAGAADKVTWTET---RTGSYG-----
Rv0223c|M.tuberculosis_H37Rv ETGQPPTIIETMHWMGSMGAMNYFAGAADKVTWTET---RTGSYG-----
Mflv_0428|M.gilvum_PYR-GCK ETGQPPTIVDMMQYGAAMSSFQFYAGAADKFTWQDI---RDGVYG-----
MAV_2101|M.avium_104 DNGMNLTAAGHIIP-VSVDMLKYYAGWVGKIHGESANLVSDGLLGTWETY
TH_4531|M.thermoresistible__bu ENGLPVSQGKAVLVPHGAGWFEAAAGWADKLEGAVVP-------TTPGET
MSMEG_6134|M.smegmatis_MC2_155 ESGLPVAR-LTGEVGRTSGQLRLFAGVLREGSWNQARIDPALPDREPLPR
Mvan_1031|M.vanbaalenii_PYR-1 ESGLPQAR-ITGELGRTTGQLRLFASVLREGSWNGARIDTALPDRTPLPR
: * .. *
MAB_3328|M.abscessus_ATCC_1997 TSSIRR-EAAGVVATITPWNYPLQMAVW--KVLPALAAGCTVVIKPSELT
MUL_2789|M.ulcerans_Agy99 TSSIRR-EAIGVVATITPWNYPLQMAVW--KVMPALAAGCTVVIKPSELT
MMAR_2610|M.marinum_M TSSIRR-EPVGPVAQVAPWNYPMMMAVW--KFAPALAAGCTVVLKPSDTT
MLBr_02573|M.leprae_Br4923 KALARY-QPLGVVLAVMPWNFPLWQTVR--FAAPALMAGNVGLLKHASNV
Mb0228c|M.bovis_AF2122/97 QSIVSR-ESVGVVGAIVAWNVPLFLAVN--KIAPALLAGCTIVLKPAAET
Rv0223c|M.tuberculosis_H37Rv QSIVSR-EPVGVVGAIVAWNVPLFLAVN--KIAPALLAGCTIVLKPAAET
Mflv_0428|M.gilvum_PYR-GCK QTLVVR-EPVGVVGAVTAWNVPFFLAAN--KLGPALLAGCTVVLKPAAET
MAV_2101|M.avium_104 HTFTQL-EPVGVVGLIIPWNGPFFVAML--KVAPALAAGCSAVLKPAEET
TH_4531|M.thermoresistible__bu VDLTLL-EPIGVVAVILTWNAPIGGWGM--CVAPPLAAGCCVILKPSELA
MSMEG_6134|M.smegmatis_MC2_155 PDIRQRSVPLGPVAVFGASNFPLAFSVAGGDTASALAAGCPVVVKAHDAH
Mvan_1031|M.vanbaalenii_PYR-1 PDLRQRHIPLGPVAVFSASNFPLAFSVAGGDTASALAAGCPVVVKGHDAH
. * * . . * *: ..* ** ::*
MAB_3328|M.abscessus_ATCC_1997 P----LTTLTLARLAAEAGLPEGVLNVVTGRGDDVGYALASHPDVDLVTF
MUL_2789|M.ulcerans_Agy99 P----LTTLTLARLAAEAGLPDGVLNVVTGLGADVGAALAAHPGVDLVTF
MMAR_2610|M.marinum_M P----ASSYWMVEKMQEI-FPPGVCNLVCG-DRDTGAALVAHPVPQLVSI
MLBr_02573|M.leprae_Br4923 P----QCALYLADVIARGGFPEDCFQTLLISANGVEAILR-DPRVAAATL
Mb0228c|M.bovis_AF2122/97 P----LTANALAEVFAEVGLPEGVLSVVPG-GIETGQALTSNPDIDMFTF
Rv0223c|M.tuberculosis_H37Rv P----LTANALAEVFAEVGLPEGVLSVVPG-GIETGQALTSNPDIDMFTF
Mflv_0428|M.gilvum_PYR-GCK P----LSVFAMAEMFVEAGLPEGVLSIVPG-GPETGQALTANPNLDKYTF
MAV_2101|M.avium_104 P----LTALKLEEIFREAGLPDGVLNVITGYGETTGAALTAHPDVDKISF
TH_4531|M.thermoresistible__bu P----FTSSFIARICEEEGMPPGVVTVLPG-GPDAGEALVRHPGVAKVSF
MSMEG_6134|M.smegmatis_MC2_155 PGTSELVGRAIAQAVTATGMPAGTFSLLYGYGPDLGSALVTDPRIKAVGF
Mvan_1031|M.vanbaalenii_PYR-1 PGTSELVARAVTDAVTTSGLPAGTFSLLFGSGPGLGIALVTDPRIKAVGF
* : :* . : * .* :
MAB_3328|M.abscessus_ATCC_1997 TGSTAVGRKVMAAAAVHGH--RTQLELGGKAPFVVFD---DADMDAAIAG
MUL_2789|M.ulcerans_Agy99 TGSTAVGRKVMAAAATHGR--RTQLELGGKAPFVVFD---DADLDAAIQG
MMAR_2610|M.marinum_M TGSTRAGMAVAASAANDLK--RAHLELGGKAPVVIFN---DADIAAAVEG
MLBr_02573|M.leprae_Br4923 TGSEPAGQAVGAIAGNEIK--PTVLELGGSDPFIVMP---SADLDAAVAT
Mb0228c|M.bovis_AF2122/97 TGSSAVGREVGRRAAEMLK--PCTLELGGKSAAIILE---DVDLAAAIPM
Rv0223c|M.tuberculosis_H37Rv TGSSAVGREVGRRAAEMLK--PCTLELGGKSAAIILE---DVDLAAAIPM
Mflv_0428|M.gilvum_PYR-GCK TGSSGVGKEIAKIAADKLK--PCTLELGGKSAAIILE---DADLDSTLPM
MAV_2101|M.avium_104 TGSTEVGRLIVKAAAGNLK--RLTLELGGKSPLIMFD---DANLDKAIMG
TH_4531|M.thermoresistible__bu TGGGPTGRKIAAACGEQLK--PVLLELGGKSANIVFP---DADLAKAAET
MSMEG_6134|M.smegmatis_MC2_155 TGSRAGGMALVAAAAARPEPIPVYAEMSAINPVFLLRGALSARAADLARA
Mvan_1031|M.vanbaalenii_PYR-1 TGSRSGGMALVSAAAARPEPIPVYAEMSSINPVFVLDGALKTRGAELGRA
**. * : .. . *:.. . .:: ..
MAB_3328|M.abscessus_ATCC_1997 AVAGAIINSGQDCTAATRALVARDL-YDDFVAGVGEVMSKVVVG-DPLDP
MUL_2789|M.ulcerans_Agy99 AVAGAVINSGQDCTAATRAIVAREL-CHDFVAGVADVMSTVVIG-DPMDP
MMAR_2610|M.marinum_M LTAAGYFNAGQDCTAATRLIIQDGI-YDEFLSAMAESVAGTRTGYDPADE
MLBr_02573|M.leprae_Br4923 AVTGRVQNNGQSCIAAKRFIAHADI-YDEFVEKFVARMETLRVG-DPTDP
Mb0228c|M.bovis_AF2122/97 MVFSGVMNAGQGCVNQTRILAPRSR-YDEIVAAVTNFVTALPVG-PPSDP
Rv0223c|M.tuberculosis_H37Rv MVFSGVMNAGQGCVNQTRILAPRSR-YDEIVAAVTNFVTALPVG-PPSDP
Mflv_0428|M.gilvum_PYR-GCK LVFSGLMNSGQACVGQTRILAPRSR-YDEVIEKLGEAVRNMAPG-LPDNP
MAV_2101|M.avium_104 AGMGLLAGSGQNCSCTSRIYVQRGI-YDQVVEGLAEFAMMLPMG-GSDDP
TH_4531|M.thermoresistible__bu AAS-VIFSAGQACTLPTRILVHESI-YDEFTAMVAGALGQIPLG-DPLDP
MSMEG_6134|M.smegmatis_MC2_155 FVGSLTMGSGQFCTNPGLVIAVDGPDLDAFVAAAAEAVAATAPT-PMLTP
Mvan_1031|M.vanbaalenii_PYR-1 FVASLTMGSGQFCTNPGLVIAVDGPGLDTFAAAARDALAGSPAT-PMLTP
. ** * . .
MAB_3328|M.abscessus_ATCC_1997 DTDIGPLISAAHRAKVSSIVDRAP-AQGGRIVTGATAPDL---PGSFYRP
MUL_2789|M.ulcerans_Agy99 DTDIGPLISLAHRDRVADILAKAP-AQGARIVVGGRIPDL---AGAFYLP
MMAR_2610|M.marinum_M DILFGPINNSNQMRHIDGLVSRAP-AH-ARILAGGKQKDC---AGFFYEP
MLBr_02573|M.leprae_Br4923 DTDVGPLATESGRAQVEQQVDAAA-AAGAVIRCGGKRPDR---PGWFYPP
Mb0228c|M.bovis_AF2122/97 AAQIGPLISEKQRTRVEALHRQG--HRGGRSVGVRRRP------------
Rv0223c|M.tuberculosis_H37Rv AAQIGPLISEKQRTRVEGYIAKGI-EEGARLVCGGGRPEGL-DNGFFIQP
Mflv_0428|M.gilvum_PYR-GCK AAMIGPLISEKQRDRVEGYIKKGI-EEGARVITGGGRPEGL-DSGWFVEP
MAV_2101|M.avium_104 NSVLGPLISEKQRQRVEGIVNDGV-AAGAKVITG-GKPMD--RKGYFYEA
TH_4531|M.thermoresistible__bu ATMMGPVISEAAVERILRTVDEATRDNGAELLTGGSRAGGELASGYYIEP
MSMEG_6134|M.smegmatis_MC2_155 GIAENFRTGVETLSGTAELIARGSDENAPAVSCRAALFGSD-VPTFLATE
Mvan_1031|M.vanbaalenii_PYR-1 TIARSYASGVEALSGAAQLVGRGAP-GTSETACHAALFSTD-AQTFLASE
. .
MAB_3328|M.abscessus_ATCC_1997 TLIADVAETSEVYR--DEIFGPVLTVRPFTDDDDALRQANDTAYGLAASA
MUL_2789|M.ulcerans_Agy99 TLLADVGEQSQAYR--EEIFGPVLTVRAHDGDDDALRQANDTDYGLAASA
MMAR_2610|M.marinum_M TLIVDLQQDDELIQ--TEIFGPVVTAQRFSDEQQALSFANGTEYGLASSV
MLBr_02573|M.leprae_Br4923 TVITDITKDMALYT--DEVFGPVASVYCATNIDDAIEIANATPFGLGSNA
Mb0228c|M.bovis_AF2122/97 ----------------------------------SRGLGQRLLYPIHERR
Rv0223c|M.tuberculosis_H37Rv TVFADVDNKMTIAQ--EEIFGPVLAIIPYDTEEDAIAIANDSVYGLAGSV
Mflv_0428|M.gilvum_PYR-GCK TVFADVDNSMTIAQ--EEIFGPVLSVIPYEDEDDAVRIANDSVYGLAGSV
MAV_2101|M.avium_104 TIITNTTPDMRLIR--EEIFGPVGCVIPFDDEDEVIAAANDTHYGLAGAI
TH_4531|M.thermoresistible__bu TLLGNVSPTSEVAQ--NEVFGPVTAAIRFSDEEEAVAIANGTPYGLAGYV
MSMEG_6134|M.smegmatis_MC2_155 ALQAEVFGASGVIVRCADDAEMLRVAAEVEGQLTATVHAEESDHAKAGEL
Mvan_1031|M.vanbaalenii_PYR-1 ALQAEVFGSSSLIVRCADFEQMRAVAEGIEGQLTATVHADDSDLDDAGRL
.:
MAB_3328|M.abscessus_ATCC_1997 WTRDVYRAQRASREIRAGCVWINDH-IPIISEMPHGGFGASGFGKDMSDY
MUL_2789|M.ulcerans_Agy99 WTRDVYRAQRASREISAGCVWINDH-IPIVSEMPHGGVAASGFGNDMSEY
MMAR_2610|M.marinum_M WTKDVDTAARMGARLDFGCVWINTH-IPLVAEMPHGGFKHSGYGKDLSMY
MLBr_02573|M.leprae_Br4923 WTQNESEQRHFIDNIVAGQVFINGM-TVSYPELPFGGIKRSGYGRELSTH
Mb0228c|M.bovis_AF2122/97 WR-----------------------------------------GKWHGQC
Rv0223c|M.tuberculosis_H37Rv WTTDVPKGIKISQQIRTGTYGIN-W-YAFDPGSPFGGYKNSGIGRENGPE
Mflv_0428|M.gilvum_PYR-GCK YTTDNDRALKIARRIRTGTYAVN-M-YAFDPCAPFGGYKNSGIGRENGWE
MAV_2101|M.avium_104 WTENLSRAHRVVNELRAGQIWVNSA-LAADPSMPISGHKQSGWGGERGRK
TH_4531|M.thermoresistible__bu HTRDVSRVLRLASALEAGTVAFNGAGAPAGPGAPFGGYKESGYGREGGKL
MSMEG_6134|M.smegmatis_MC2_155 LEILELKAGRILFNGWPTGVEVGHAMVHGGPFPSTSDSRTTSVGARAIER
Mvan_1031|M.vanbaalenii_PYR-1 LPLLELKAGRILFGGWPTGVEVCHAMVHGGPFPATSDSRSTSVGSQAIER
*
MAB_3328|M.abscessus_ATCC_1997 SLEEYLTVKHVMSDITGTADKDWHRTIFAKR-------
MUL_2789|M.ulcerans_Agy99 SFDEYLTIKHVMSDITGVAEKPWHRTIFSTRI------
MMAR_2610|M.marinum_M GFEDYTRIKHVM-----------HYHGFEG--------
MLBr_02573|M.leprae_Br4923 GIREFCNIKTVWIA------------------------
Mb0228c|M.bovis_AF2122/97 G-------------------------------------
Rv0223c|M.tuberculosis_H37Rv GVEHFTQQKSVLLPMGYTVA------------------
Mflv_0428|M.gilvum_PYR-GCK GIEAYCEQKSILLPFGYTPPAS----------------
MAV_2101|M.avium_104 GIEAYFNTKAVYIGL-----------------------
TH_4531|M.thermoresistible__bu GVLEFVQHKTVSVALS----------------------
MSMEG_6134|M.smegmatis_MC2_155 FLRPVCYQNAPKSLLPSAIADGNPDGMWRTVDGQLTKD
Mvan_1031|M.vanbaalenii_PYR-1 YLRPVCYQDVPAPLLPSAIAEGNPEKLWRRVDGRLTQD