For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LHTVETPDARRSDSPVEYRWYAGGTWRPADEDWFDDADPWSGEVYARVARCGRQEAAAAVQAAADAYPAW SQTTPREKARLFRAAAAVVARRRDDLAVTLARETGTTKVNADFQITLTIELIEQAAGWVYLPAGEVLRSD FADTSQTVVRRPLGVVASFTPWNGATILSWRAALSPIAAGNTVVVKPSELAPVSAGLQLAEIMEEAGFPA GVVNVVTHGPGEAGPIADEFFDNPLVRCINFIGSVPTGRMLAERAGATLKRSVMELGGYNPLIILEDVDL DYAVRVAAFSAFFHQGQICLNARKILVHRDIYDEFVARLVAKAESLTSGDPLHADTFIGPLITPAAVELV HARVNDAVAKGARLLTGGTYDGPVYAPTVLVDVPDDAVVSSEETFGPVVVVHPVDTAEEAIEIANRPLYG LSSAILSGDTHRATKLAESLRSGVVRVNLPTIDDEIQAPIGGVRDSGWGRSGPWALQDFTDLVAIAVQSG QRDLPIS
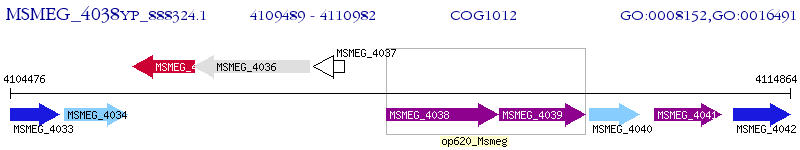
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4038 | - | - | 100% (497) | vanillin dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2488 | - | 4e-74 | 37.15% (471) | [NADP+] succinate-semialdehyde dehydrogenase [Mycobacterium |
| M. smegmatis MC2 155 | MSMEG_4165 | - | 4e-71 | 35.90% (454) | [NAD+] benzaldehyde dehydrogenase [Mycobacterium smegmatis str. |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0791 | aldA | 5e-54 | 33.05% (472) | aldehyde dehydrogenase NAD dependant AldA |
| M. gilvum PYR-GCK | Mflv_1858 | - | 3e-63 | 32.56% (473) | succinate-semialdehyde dehydrogenase (NAD(P)(+)) |
| M. tuberculosis H37Rv | Rv0223c | - | 8e-63 | 33.74% (495) | aldehyde dehydrogenase |
| M. leprae Br4923 | MLBr_02573 | gabD1 | 1e-49 | 30.63% (444) | succinic semialdehyde dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3328 | - | 4e-66 | 34.74% (449) | gamma-aminobutyraldehyde dehydrogenase |
| M. marinum M | MMAR_2875 | - | 2e-63 | 36.15% (473) | aldehyde dehydrogenase |
| M. avium 104 | MAV_2101 | - | 3e-58 | 34.68% (444) | aldehyde dehydrogenase |
| M. thermoresistible (build 8) | TH_4137 | - | 1e-79 | 37.87% (478) | PUTATIVE putative [NADP+] succinate-semialdehyde dehydrogenase |
| M. ulcerans Agy99 | MUL_2877 | - | 2e-62 | 35.94% (473) | aldehyde dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0014 | - | 4e-67 | 36.27% (488) | betaine-aldehyde dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2875|M.marinum_M --------MTTVAVPHHAMLIGGES-VDS-DRRLSLVDPGDESPVATVAC
MUL_2877|M.ulcerans_Agy99 --------MTTVAVPHHAMLIGGES-VDS-DRRLSLVDPGDESPVATVAC
Mvan_0014|M.vanbaalenii_PYR-1 ----------MSDIPHYRMYIDGAW-RDA-ADTIEVRSPATGELVATVAY
Mb0791|M.bovis_AF2122/97 --------MALWGDGISALLIDGKL-SDGRAGTFPTVNPATEEVLGVAAD
Rv0223c|M.tuberculosis_H37Rv --------MSDSATEYDKLFIGGKWTKPSTSDVIEVRCPATGEYVGKVPM
MAV_2101|M.avium_104 -----------------------------MSTTIPVFNPSTEEQIAEVPD
MAB_3328|M.abscessus_ATCC_1997 -------------MSVPSSWINGRP-VATHGDTHRVINPATGEAVADLKL
MLBr_02573|M.leprae_Br4923 -------------------------------MSIATINPATGETVKTFTP
Mflv_1858|M.gilvum_PYR-GCK --MDNSAVEALLASVPTGLWIGGEEREGTS--TFDVLDPSDDTVLTSVAD
TH_4137|M.thermoresistible__bu ------------------MYIDGEWQSAASGKTFASINPANGAVLDSVPD
MSMEG_4038|M.smegmatis_MC2_155 MHTVETPDARRSDSPVEYRWYAGGTWRPADEDWFDDADPWSGEVYARVAR
*
MMAR_2875|M.marinum_M GDASHVDAAVAAAKASFEDGSWSRATPQERAAVMRRIVAAADQVTDELIE
MUL_2877|M.ulcerans_Agy99 GDASHVDAAVAAAKASFEDGSWSRATPQERAAVMRRIVAAADQVTDELIE
Mvan_0014|M.vanbaalenii_PYR-1 GGVTTVDDAVAAAKAADEAGVWRNTSAQQRADVLDAIADNLAARTDELTA
Mb0791|M.bovis_AF2122/97 ADAEDMGRAIEAARRAFDSTDWSRNT-ELRVRCVRQLRDAMQQHVEELRE
Rv0223c|M.tuberculosis_H37Rv AAAADVDAAVAAARAAFDNGPWPSTPPHERAAVIAAAVKMLAERKDLFTK
MAV_2101|M.avium_104 SDQAAVDAAVARARETFESGVWRKAPASHRANVLFRAARIIEERKDELAA
MAB_3328|M.abscessus_ATCC_1997 ATTADVDAAVAAARAAG--PAWASSTPVDRSSVLAKLAAVLSANADTLVA
MLBr_02573|M.leprae_Br4923 TTQDEVEDAIARAYARFD--NYRHTTFTQRAKWANATADLLEAEANQSAA
Mflv_1858|M.gilvum_PYR-GCK ATADDARAALDAACAVQA--EWAGTAPRKRGEILRSVFETITARADDIAA
TH_4137|M.thermoresistible__bu GEREDARRAIDAAAAALP--DWRSRTAYERAEVLVEAHRIMLERRDDLAE
MSMEG_4038|M.smegmatis_MC2_155 CGRQEAAAAVQAAADAYP--AWSQTTPREKARLFRAAAAVVARRRDDLAV
*: * : . : :
MMAR_2875|M.marinum_M LEMACNGATIRLATGFHIGYALSHFSYFADLAESYAYQR-PAPIETFP--
MUL_2877|M.ulcerans_Agy99 LEMACNGATIRLATGFHIGHALSHFSYFADLAESYAYQR-PAPIETFP--
Mvan_0014|M.vanbaalenii_PYR-1 LQVAENGATVRAAGAFLIGYAIAHLRYFAGLARTYAFQS-SGPLMEAP--
Mb0791|M.bovis_AF2122/97 LTISEVGAPRMLTASAQLEGPVGDLSFAADTAESYPWKQDLGEASPLG--
Rv0223c|M.tuberculosis_H37Rv LLAAETGQPPTIIETMHWMGSMGAMNYFAGAADKVTWTETRTGSYG----
MAV_2101|M.avium_104 LESQDNGMNLTAAGHIIPV-SVDMLKYYAGWVGKIHGESANLVSDGLLGT
MAB_3328|M.abscessus_ATCC_1997 DEVAQTGKPVRLASEFDVPGSIDNVEFFAGAARHLEGKATAEYSADHT--
MLBr_02573|M.leprae_Br4923 LMTLEMGKTLQSAKDEAMKCAKGFRYYAEKAETLLADEPAEAAAVGAS--
Mflv_1858|M.gilvum_PYR-GCK LMTLEMGKVLAESK-GEVTYGAEFFRWFAEEAVRIDGRYTAAPAGTGR--
TH_4137|M.thermoresistible__bu LMTREQGKPLRMAR-TEVGYAADFLLWFAEEAKRVYGEVIPSARADQR--
MSMEG_4038|M.smegmatis_MC2_155 TLARETG-TTKVNADFQITLTIELIEQAAGWVYLPAGEVLRSDFADTS--
*
MMAR_2875|M.marinum_M -VLSQSVVHREPIGVVGAITPWNFPLLLTLWKVGPAIAAGNSVVVKPDEH
MUL_2877|M.ulcerans_Agy99 -VLSQSVVHREPIGVVGAITPWNFPLLLTLWKVGPAIAAGNSVVVKPDEH
Mvan_0014|M.vanbaalenii_PYR-1 -TLAAGMIVRDPVGVCAGMIPWNFPLLLAVWKLGPALAAGNTVVLKPDDQ
Mb0791|M.bovis_AF2122/97 -IATRRTLAREAVGVVGAITPWNFPHQINLAKLGPALAAGNTVVLKPAPD
Rv0223c|M.tuberculosis_H37Rv ----QSIVSREPVGVVGAIVAWNVPLFLAVNKIAPALLAGCTIVLKPAAE
MAV_2101|M.avium_104 WETYHTFTQLEPVGVVGLIIPWNGPFFVAMLKVAPALAAGCSAVLKPAEE
MAB_3328|M.abscessus_ATCC_1997 -----SSIRREAAGVVATITPWNYPLQMAVWKVLPALAAGCTVVIKPSEL
MLBr_02573|M.leprae_Br4923 ----KALARYQPLGVVLAVMPWNFPLWQTVRFAAPALMAGNVGLLKHASN
Mflv_1858|M.gilvum_PYR-GCK -----ILVTKQAVGPCYAITPWNFPLAMGTRKMGPAFAAGCTMIVKPAQE
TH_4137|M.thermoresistible__bu -----FIVLAQPVGVCAAITPWNYPISMLTRKLAPALAAGCTSVLKPAEQ
MSMEG_4038|M.smegmatis_MC2_155 -----QTVVRRPLGVVASFTPWNGATILSWRAALSPIAAGNTVVVKPSEL
. * . .** . ..: ** ::*
MMAR_2875|M.marinum_M TPLSILEFAR-IAERSGLPPGVINVVPGE---GPEAGARLASHPDVGKIA
MUL_2877|M.ulcerans_Agy99 TPLSILEFAR-IAERSGLPPGVINVVPGE---GPEAGARLASHPDVGKIA
Mvan_0014|M.vanbaalenii_PYR-1 TPLTLLELAR-AADEVGLPAGVLNVVTGA---GPTVGARLAEHPDVRKVA
Mb0791|M.bovis_AF2122/97 TPWCAAALGEIIVEHTDFPPGVVNIVTSS---SHALGALLAKDPRVDMIS
Rv0223c|M.tuberculosis_H37Rv TPLTANALAE-VFAEVGLPEGVLSVVPG----GIETGQALTSNPDIDMFT
MAV_2101|M.avium_104 TPLTALKLEE-IFREAGLPDGVLNVITGY---GETTGAALTAHPDVDKIS
MAB_3328|M.abscessus_ATCC_1997 TPLTTLTLAR-LAAEAGLPEGVLNVVTGR---GDDVGYALASHPDVDLVT
MLBr_02573|M.leprae_Br4923 VPQCALYLAD-VIARGGFPEDCFQTLLIS---ANGVEAILR-DPRVAAAT
Mflv_1858|M.gilvum_PYR-GCK TPLTMLLLAK-LMADAGLPKGVLSVLPTTS--PGPVTETLIDDGRLRKLT
TH_4137|M.thermoresistible__bu TPLCAVETFK-VFADAGVPAGVVNLVTCSD--PEPVADELLGDPRVRKIS
MSMEG_4038|M.smegmatis_MC2_155 APVSAGLQLAEIMEEAGFPAGVVNVVTHGPGEAGPIADEFFDNPLVRCIN
.* ..* . .. : : . :
MMAR_2875|M.marinum_M FTGSTAVGREIMRLASGTVKQVTLELGGKGPTIVLDDADLEMAVEGVLYG
MUL_2877|M.ulcerans_Agy99 FTGSTAVGREIMRLASGTVKQVTLELGGKGPTIVLDDADLEMAVEGVLYG
Mvan_0014|M.vanbaalenii_PYR-1 FTGSTEVGKSVMRAAADTVKKVTLELGGKGASIVLDDADLDLAVDGSLFA
Mb0791|M.bovis_AF2122/97 FTGSTATGRAVMADAAATIKKVFLELGGKSAFVVLDDADLAAASAVSAFS
Rv0223c|M.tuberculosis_H37Rv FTGSSAVGREVGRRAAEMLKPCTLELGGKSAAIILEDVDLAAAIPMMVFS
MAV_2101|M.avium_104 FTGSTEVGRLIVKAAAGNLKRLTLELGGKSPLIMFDDANLDKAIMGAGMG
MAB_3328|M.abscessus_ATCC_1997 FTGSTAVGRKVMAAAAVHGHRTQLELGGKAPFVVFDDADMDAAIAGAVAG
MLBr_02573|M.leprae_Br4923 LTGSEPAGQAVGAIAGNEIKPTVLELGGSDPFIVMPSADLDAAVATAVTG
Mflv_1858|M.gilvum_PYR-GCK FTGSTGVGKALVKQSADKLLRTSMELGGNAPFVVFDDADVDAAVDGAILA
TH_4137|M.thermoresistible__bu FTGSTEVGRMIASRAGATMKRVSMELGGHAPFLVFPDADPEHAAKGGAAV
MSMEG_4038|M.smegmatis_MC2_155 FIGSVPTGRMLAERAGATLKRSVMELGGYNPLIILEDVDLDYAVRVAAFS
: ** .*: : :. :**** . ::: ..: *
MMAR_2875|M.marinum_M CFAYSGQVCESGTRALVPTTLYQEFVDRLVKRAATIVVGPTRDWDTDLGP
MUL_2877|M.ulcerans_Agy99 CFAYSGQVCESGTRALVPTTLYQEFVDRLVKRAATIVVGPTRDWDTDLGP
Mvan_0014|M.vanbaalenii_PYR-1 FLLMSGQACESGTRLLVHESIHDEFVRRMVARAETLVMGDPMSLASDLGP
Mb0791|M.bovis_AF2122/97 ACMHAGQGCAITTRLVVPRARYEEAVAIAAATMSSIRPGDPNDPGTVCGP
Rv0223c|M.tuberculosis_H37Rv GVMNAGQGCVNQTRILAPRSRYDEIVAAVTNFVTALPVGPPSDPAAQIGP
MAV_2101|M.avium_104 LLAGSGQNCSCTSRIYVQRGIYDQVVEGLAEFAMMLPMGGSDDPNSVLGP
MAB_3328|M.abscessus_ATCC_1997 AIINSGQDCTAATRALVARDLYDDFVAGVGEVMSKVVVGDPLDPDTDIGP
MLBr_02573|M.leprae_Br4923 RVQNNGQSCIAAKRFIAHADIYDEFVEKFVARMETLRVGDPTDPDTDVGP
Mflv_1858|M.gilvum_PYR-GCK KMRNGGEACTAANRIHVANAVREEFTDKFVKRMSEFTLGKGLDEKSTLGP
TH_4137|M.thermoresistible__bu KFLNTGQACICPNRFIVHRSIAEPFIATLRARIEKLVPGDGLAAGTTVGP
MSMEG_4038|M.smegmatis_MC2_155 AFFHQGQICLNARKILVHRDIYDEFVARLVAKAESLTSGDPLHADTFIGP
*: * : . : . * : **
MMAR_2875|M.marinum_M VIDAAQRTRILNYIRGAQTDGATVALGGEALTGGMFDKGFWVAPTILTGV
MUL_2877|M.ulcerans_Agy99 VIDAAQRTRILNYIRGAQTDGATVALGGEALTGGMFDKGFWVAPTILTGV
Mvan_0014|M.vanbaalenii_PYR-1 LVSAKQKARVEKYIALGQEEGCKLAYQGTVPTDPALAQGHWVPPTILTGA
Mb0791|M.bovis_AF2122/97 LISARQRDRVQGYLDLAVAEGGRFACGGARPAD--REVGFYIEPTVIAGL
Rv0223c|M.tuberculosis_H37Rv LISEKQRTRVEGYIAKGIEEGARLVCGGGRPEG--LDNGFFIQPTVFADV
MAV_2101|M.avium_104 LISEKQRQRVEGIVNDGVAAGAKVITGG-KPMD--RK-GYFYEATIITNT
MAB_3328|M.abscessus_ATCC_1997 LISAAHRAKVSSIVDRAPAQGGRIVTGATAPDLP----GSFYRPTLIADV
MLBr_02573|M.leprae_Br4923 LATESGRAQVEQQVDAAAAAGAVIRCGGKRPDRP----GWFYPPTVITDI
Mflv_1858|M.gilvum_PYR-GCK LINAKQVATVTELVSDAVSRGATVAVGGVAPGGP----GNFYPATVLTDV
TH_4137|M.thermoresistible__bu LIDEVALKKMQRQVDDAVDKGAKLELGGSRLQGPAFDNGHFYAPTILTGV
MSMEG_4038|M.smegmatis_MC2_155 LITPAAVELVHARVNDAVAKGARLLTGGTYDGP-------VYAPTVLVDV
: : : . * . . .*::..
MMAR_2875|M.marinum_M HNDMAIAREEVFGPVLVVIGYDGDEQAIAIANDSDYGLAASIWTSDNARG
MUL_2877|M.ulcerans_Agy99 HNDMAIAREEVFGPVLVVSGYDGDEQAIAIANDSDYGLAASIWTSDNARG
Mvan_0014|M.vanbaalenii_PYR-1 TNDMRIAREEIFGPVLVVLTYGDDDEAVAIANDSEYGLSAGVWSADRERA
Mb0791|M.bovis_AF2122/97 TNDARVAREEIFGPVLTVIAHDGDDDAVRIANDSPYGLSGTVYGADPQRA
Rv0223c|M.tuberculosis_H37Rv DNKMTIAQEEIFGPVLAIIPYDTEEDAIAIANDSVYGLAGSVWTTDVPKG
MAV_2101|M.avium_104 TPDMRLIREEIFGPVGCVIPFDDEDEVIAAANDTHYGLAGAIWTENLSRA
MAB_3328|M.abscessus_ATCC_1997 AETSEVYRDEIFGPVLTVRPFTDDDDALRQANDTAYGLAASAWTRDVYRA
MLBr_02573|M.leprae_Br4923 TKDMALYTDEVFGPVASVYCATNIDDAIEIANATPFGLGSNAWTQNESEQ
Mflv_1858|M.gilvum_PYR-GCK PADARILKEEVFGPVAPIAGFDTEEEGIAAANDTEYGLAAYVYTQSLDRA
TH_4137|M.thermoresistible__bu TPDMLICSEETFGPIAPVMVFDDEDEAIAMANSTEYGLAGYIYTRDISRA
MSMEG_4038|M.smegmatis_MC2_155 PDDAVVSSEETFGPVVVVHPVDTAEEAIEIANRPLYGLSSAILSGDTHRA
: :* ***: : :: : ** . :**.. . .
MMAR_2875|M.marinum_M LAMAERIQSGSVWINDAHQINCR-LPFGGYKQSGVGRELGPDALDAYTEV
MUL_2877|M.ulcerans_Agy99 LAMAERIRSGSVWINDAHQINCR-LLFGGYKQSGVGRELGPDTLDAYTEV
Mvan_0014|M.vanbaalenii_PYR-1 LGIARRLQSGTVWVNDWHMINAM-YPFGGVKQSGLGRELGPDALDEYTEP
Mb0791|M.bovis_AF2122/97 ARIASRLRVGTVNVNGGVWYCAD-APFGGYKQSGIGREMGLLGFEEYLEA
Rv0223c|M.tuberculosis_H37Rv IKISQQIRTGTYGIN-WYAFDPG-SPFGGYKNSGIGRENGPEGVEHFTQQ
MAV_2101|M.avium_104 HRVVNELRAGQIWVNSALAADPS-MPISGHKQSGWGGERGRKGIEAYFNT
MAB_3328|M.abscessus_ATCC_1997 QRASREIRAGCVWINDHIPIISE-MPHGGFGASGFGKDMSDYSLEEYLTV
MLBr_02573|M.leprae_Br4923 RHFIDNIVAGQVFINGMTVSYPE-LPFGGIKRSGYGRELSTHGIREFCNI
Mflv_1858|M.gilvum_PYR-GCK LRVAEGIESGMVGVNRGVISDPA-APFGGIKESGFGREGGTEGIEEYLDT
TH_4137|M.thermoresistible__bu IRVGEALDFGMIGINDINPTSAA-VPFGGIKQSGLGREGARAGIEEYLDT
MSMEG_4038|M.smegmatis_MC2_155 TKLAESLRSGVVRVNLPTIDDEIQAPIGGVRDSGWGRS-GPWALQDFTDL
: * :* .* ** * . . . :
MMAR_2875|M.marinum_M KTIHLDLSGGREAKPYDVLLGHPETG
MUL_2877|M.ulcerans_Agy99 KTIHLDLSGGREAKPYDVLLGHPETG
Mvan_0014|M.vanbaalenii_PYR-1 KFIHVDMTDDRRKHVYPVVISAAAQG
Mb0791|M.bovis_AF2122/97 KLIATAAN------------------
Rv0223c|M.tuberculosis_H37Rv KSVLLPMGYTVA--------------
MAV_2101|M.avium_104 KAVYIGL-------------------
MAB_3328|M.abscessus_ATCC_1997 KHVMSDITGTADKDWHRTIFAKR---
MLBr_02573|M.leprae_Br4923 KTVWIA--------------------
Mflv_1858|M.gilvum_PYR-GCK KYIALTK-------------------
TH_4137|M.thermoresistible__bu KVLGIAL-------------------
MSMEG_4038|M.smegmatis_MC2_155 VAIAVQSGQRDLPIS-----------
: