For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GFMTTTLASTHPVETVHGHRVPTGLFIGGDWRTTDSTFTVLDPATTGHIADVSDAGAAEALAALDAAHAV RDEWRRVTPRARAGLFGEAYRLLRSRAEAFAAVMTAESGKPLAESRAEFNLSAEFFLWYTEQIAHLHGTY APASHGGYRVVTTHQPVGPALLITPWNFPMLMIARKAGAALAAGCPVIVKSAKETPLTCALFVETLREAG FPAGVVNLVHTTSSSAVSAALMADPRLRKISFTGSTGVGSTLLAQAAPGILNASMELGGDGPFIVLEDAD VELAAKQAVVCKFRNAGQACVAANRIIVHRAVAEEFTKRFVALTEQLRVGNGFDDGVDVGPIISARQLDG VSELVDTLRGLDSELLTGGNALPGDGFFFAPTVFRVNGRPTELCSRELFAPVATIYQVESTSEAIEFAND TRYGLAAYVFTRDLSRAVAVGERLDFGMVGINRGIMADPAAAFGGVKASGLGREGGHDAIYDYLEPKYLA LTVDEEAGLA*
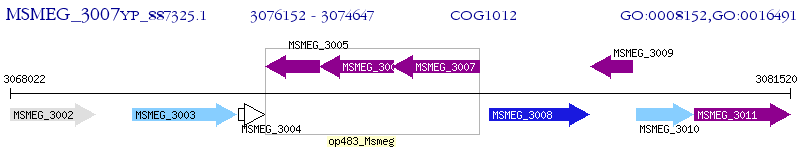
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3007 | - | - | 100% (501) | succinate-semialdehyde dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5538 | - | e-121 | 47.46% (472) | [NADP+] succinate-semialdehyde dehydrogenase [Mycobacterium |
| M. smegmatis MC2 155 | MSMEG_2488 | - | e-118 | 47.00% (483) | [NADP+] succinate-semialdehyde dehydrogenase [Mycobacterium |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2883c | aldC | 1e-62 | 33.62% (461) | aldehyde dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1858 | - | 1e-123 | 48.09% (472) | succinate-semialdehyde dehydrogenase (NAD(P)(+)) |
| M. tuberculosis H37Rv | Rv2858c | aldC | 1e-62 | 33.62% (461) | aldehyde dehydrogenase |
| M. leprae Br4923 | MLBr_02573 | gabD1 | 2e-58 | 32.16% (454) | succinic semialdehyde dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4208 | - | 1e-120 | 46.30% (473) | putative succinate-semialdehyde dehydrogenase |
| M. marinum M | MMAR_3556 | - | 4e-65 | 32.84% (475) | aldehyde dehydrogenase NAD dependent |
| M. avium 104 | MAV_1897 | - | 2e-73 | 34.97% (489) | betaine-aldehyde dehydrogenase |
| M. thermoresistible (build 8) | TH_4137 | - | 1e-103 | 42.11% (475) | PUTATIVE putative [NADP+] succinate-semialdehyde dehydrogenase |
| M. ulcerans Agy99 | MUL_2789 | - | 4e-64 | 32.63% (475) | gamma-aminobutyraldehyde dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_4876 | - | 1e-123 | 48.09% (472) | succinate-semialdehyde dehydrogenase (NAD(P)(+)) |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2883c|M.bovis_AF2122/97 --MSTTQ-----LINPATEEVL----------------------------
Rv2858c|M.tuberculosis_H37Rv --MSTTQ-----LINPATEEVL----------------------------
MAV_1897|M.avium_104 --MTDTT-----ISNGVTEQLVHFDLVIGGESVAAESGRTYDSIDPYTAL
MMAR_3556|M.marinum_M --MTTTVSVAGSWINGAAHSTG---------------GRQHAVINPATQE
MUL_2789|M.ulcerans_Agy99 --MTTTVSVAGSWINGAAHSTG---------------GRQHAVINPATQE
Mflv_1858|M.gilvum_PYR-GCK ---------MDNSAVEALLASVPTGLWIGGEEREGTS--TFDVLDPSDDT
Mvan_4876|M.vanbaalenii_PYR-1 ---------MDTS---ALLKSVPTGLWIGGEERQGTS--TFSVLDPSDDE
MAB_4208|M.abscessus_ATCC_1997 --------MTDHS---TLIDAIPTDLWIGGKQVPSSTGTRFPVHNPATGA
MSMEG_3007|M.smegmatis_MC2_155 MGFMTTTLASTHPVETVHGHRVPTGLFIGGDWRTTDS--TFTVLDPATTG
TH_4137|M.thermoresistible__bu -------------------------MYIDGEWQSAASGKTFASINPANGA
MLBr_02573|M.leprae_Br4923 --------------------------------------MSIATINPATGE
Mb2883c|M.bovis_AF2122/97 --ASVDHTDANAVDDAVQRARAAQR-RWARLAPAQRAAGLRAFAAAVQAH
Rv2858c|M.tuberculosis_H37Rv --ASVDHTDANAVDDAVQRARAAQR-RWARLAPAQRAAGLRAFAAAVQAH
MAV_1897|M.avium_104 PWARVPDCGPADVDRAVAAARAALRGPWGQLTGTARGKLLWRLGELIARE
MMAR_3556|M.marinum_M TIAELALATPADVDRAVVAARRALP-QWAGVTPAERSAVLAKLADLLEQR
MUL_2789|M.ulcerans_Agy99 TIAELALATPADVDHAVVAARRALP-QWAGVTTAERSAVLAELADLLEQR
Mflv_1858|M.gilvum_PYR-GCK VLTSVADATADDARAALDAACAVQA-EWAGTAPRKRGEILRSVFETITAR
Mvan_4876|M.vanbaalenii_PYR-1 VLATVADATADDARDALDAACAVQG-EWAATPARKRGEILRSVFETITAR
MAB_4208|M.abscessus_ATCC_1997 VLTTVADAAASDGAIALDEATAVQR-SWAATAPRERAEILRRAWELVIAR
MSMEG_3007|M.smegmatis_MC2_155 HIADVSDAGAAEALAALDAAHAVRD-EWRRVTPRARAGLFGEAYRLLRSR
TH_4137|M.thermoresistible__bu VLDSVPDGEREDARRAIDAAAAALP-DWRSRTAYERAEVLVEAHRIMLER
MLBr_02573|M.leprae_Br4923 TVKTFTPTTQDEVEDAIARAYARFD-NYRHTTFTQRAKWANATADLLEAE
. *: * : . *. : .
Mb2883c|M.bovis_AF2122/97 LDELAALEVANSGH-PIVSAEWEAGHVRDVLAFYAASPERLSG--RQIPV
Rv2858c|M.tuberculosis_H37Rv LDELAALEVANSGH-PIVSAEWEAGHVRDVLAFYAASPERLSG--RQIPV
MAV_1897|M.avium_104 AESLAELEVRDGGK-LVREMIGQMKALPDYYIYYAGLADKLQG--EVVPV
MMAR_3556|M.marinum_M APELIAEEVCQTGKPLRLAEEFDVPGSVDNIRFFAGAARRLPG--QATAE
MUL_2789|M.ulcerans_Agy99 APELIAEEVCQTGKPLRLAEEFDVPGSVDNIRFFAGAARRLPG--QATAE
Mflv_1858|M.gilvum_PYR-GCK ADDIAALMTLEMGK-VLAESKGEVTYGAEFFRWFAEEAVRIDG--RYTAA
Mvan_4876|M.vanbaalenii_PYR-1 ADDIAALMTLEMGK-VFAESKGEVTYGAEFFRWFAEEAVRIEG--RYTPA
MAB_4208|M.abscessus_ATCC_1997 RDDFALAMTLEMGK-PLAESQGEVAYGGEFLRWFSEEAVRING--RYTSS
MSMEG_3007|M.smegmatis_MC2_155 AEAFAAVMTAESGK-PLAESRAEFNLSAEFFLWYTEQIAHLHG--TYAPA
TH_4137|M.thermoresistible__bu RDDLAELMTREQGK-PLRMARTEVGYAADFLLWFAEEAKRVYG--EVIPS
MLBr_02573|M.leprae_Br4923 ANQSAALMTLEMGK-TLQSAKDEAMKCAKGFRYYAEKAETLLADEPAEAA
. : *: : . ::: : . .
Mb2883c|M.bovis_AF2122/97 AGG--VDVTFNEPMGVVGVITPWNFPMVIASWAIAPALAAGNAVLVKPAE
Rv2858c|M.tuberculosis_H37Rv AGG--VDVTFNEPMGVVGVITPWNFPMVIASWAIAPALAAGNAVLVKPAE
MAV_1897|M.avium_104 DKPNFFVYTRHEPVGVVGAITPWNSPLLLLTWKLAAGLAAGCTFVVKPSD
MMAR_3556|M.marinum_M YSADHTSSIRREAIGVVATITPWNYPLQMAVWKVMPALAAGCTVVIKPSE
MUL_2789|M.ulcerans_Agy99 YSADHTSSIRREAIGVVATITPWNYPLQMAVWKVMPALAAGCTVVIKPSE
Mflv_1858|M.gilvum_PYR-GCK PAGTGRILVTKQAVGPCYAITPWNFPLAMGTRKMGPAFAAGCTMIVKPAQ
Mvan_4876|M.vanbaalenii_PYR-1 PAGTGRILVTKQAVGPCYAITPWNFPLAMGTRKMGPAFAAGCTMIVKPAQ
MAB_4208|M.abscessus_ATCC_1997 PTGAGRILVAKQPIGPTLAITPWNFPLAMGTRKIGPALAAGCTMIVKPAA
MSMEG_3007|M.smegmatis_MC2_155 SHGGYRVVTTHQPVGPALLITPWNFPMLMIARKAGAALAAGCPVIVKSAK
TH_4137|M.thermoresistible__bu ARADQRFIVLAQPVGVCAAITPWNYPISMLTRKLAPALAAGCTSVLKPAE
MLBr_02573|M.leprae_Br4923 AVGASKALARYQPLGVVLAVMPWNFPLWQTVRFAAPALMAGNVGLLKHAS
:.:* : *** *: ..: ** ::* :
Mb2883c|M.bovis_AF2122/97 LTPLTTMRLGELAVEAGLDEDLLQVLPG-KGTVVGERFVTHPDIRKIVFT
Rv2858c|M.tuberculosis_H37Rv LTPLTTMRLGELAVEAGLDEDLLQVLPG-KGTVVGERFVTHPDIRKIVFT
MAV_1897|M.avium_104 HTPASTLAFAKLFAEAGFPPGVINVVTG-WGPQTGAALAAHPDVDKIAFT
MMAR_3556|M.marinum_M LTPLTTLTLARLAAEAGLPDGVLNVVTG-LGADVGAALAAHPGVDLVTFT
MUL_2789|M.ulcerans_Agy99 LTPLTTLTLARLAAEAGLPDGVLNVVTG-LGADVGAALAAHPGVDLVTFT
Mflv_1858|M.gilvum_PYR-GCK ETPLTMLLLAKLMADAGLPKGVLSVLPTTSPGPVTETLIDDGRLRKLTFT
Mvan_4876|M.vanbaalenii_PYR-1 ETPLTMLLLAKLMAEAGLPKGVLSVLPTSNPGAVTEALINDGRLRKLTFT
MAB_4208|M.abscessus_ATCC_1997 ETPLTMLLLGQVFADAGLPAGVLSILPTTNASALTGPLIDDPRLRKLTFT
MSMEG_3007|M.smegmatis_MC2_155 ETPLTCALFVETLREAGFPAGVVNLVHTTSSSAVSAALMADPRLRKISFT
TH_4137|M.thermoresistible__bu QTPLCAVETFKVFADAGVPAGVVNLVTCSDPEPVADELLGDPRVRKISFT
MLBr_02573|M.leprae_Br4923 NVPQCALYLADVIARGGFPEDCFQTLLISANG--VEAILRDPRVAAATLT
.* .*. . .. : : . : :*
Mb2883c|M.bovis_AF2122/97 GSTEVGKRVMAGAAAQVKRVTLELGGKSANIVFHDCDLERAATTAPAGVF
Rv2858c|M.tuberculosis_H37Rv GSTEVGKRVMAGAAAQVKRVTLELGGKSANIVFHDCDLERAATTAPAGVF
MAV_1897|M.avium_104 GSTATGISVGKAAIDNMTRFTLELGGKSAQVVFDDADLDAAANGVIAGVF
MMAR_3556|M.marinum_M GSTAVGRKVMAAAATHGRRTQLELGGKAPFVVFDDADLDAAIQGAVAGAL
MUL_2789|M.ulcerans_Agy99 GSTAVGRKVMAAAATHGRRTQLELGGKAPFVVFDDADLDAAIQGAVAGAV
Mflv_1858|M.gilvum_PYR-GCK GSTGVGKALVKQSADKLLRTSMELGGNAPFVVFDDADVDAAVDGAILAKM
Mvan_4876|M.vanbaalenii_PYR-1 GSTGVGKALVKQSADKLLRTSMELGGNAPFIVFDDADVDAAVDGAILAKM
MAB_4208|M.abscessus_ATCC_1997 GSTGVGKILLSQCATRVLRSSMELGGNAPFVVFDDADVDAAVDGAMAAKM
MSMEG_3007|M.smegmatis_MC2_155 GSTGVGSTLLAQAAPGILNASMELGGDGPFIVLEDADVELAAKQAVVCKF
TH_4137|M.thermoresistible__bu GSTEVGRMIASRAGATMKRVSMELGGHAPFLVFPDADPEHAAKGGAAVKF
MLBr_02573|M.leprae_Br4923 GSEPAGQAVGAIAGNEIKPTVLELGGSDPFIVMPSADLDAAVATAVTGRV
** .* : . :**** . :*: ..* : * .
Mb2883c|M.bovis_AF2122/97 DNAGQDCCARSRILVQRSVYDRFMELLEPAVHSIVVGDPGSRATEMGPLV
Rv2858c|M.tuberculosis_H37Rv DNAGQDCCARSRILVQRSVYDRFMELLEPAVHSIVVGDPGSRATEMGPLV
MAV_1897|M.avium_104 AATGQTCLAGSRLLVHESVADALVERIVARASTIKLGDPKDPATEMGPVS
MMAR_3556|M.marinum_M INSGQDCTAATRAIVARELYHDFVAGVADVMSTVVIGDPMDPDTDIGPLI
MUL_2789|M.ulcerans_Agy99 INSGQDCTAATRAIVARELCHDFVAGVADVMSTVVIGDPMDPDTDIGPLI
Mflv_1858|M.gilvum_PYR-GCK RNGGEACTAANRIHVANAVREEFTDKFVKRMSEFTLGKGLDEKSTLGPLI
Mvan_4876|M.vanbaalenii_PYR-1 RNGGEACTAANRIHVANAVREEFTEKFVKRMSEFTLGKGLDEKSTLGPLI
MAB_4208|M.abscessus_ATCC_1997 RNIGEACTAANRFHVDNAVRAEFTEKLTARMGALTIGPGDENGVQVGPLI
MSMEG_3007|M.smegmatis_MC2_155 RNAGQACVAANRIIVHRAVAEEFTKRFVALTEQLRVGNGFDDGVDVGPII
TH_4137|M.thermoresistible__bu LNTGQACICPNRFIVHRSIAEPFIATLRARIEKLVPGDGLAAGTTVGPLI
MLBr_02573|M.leprae_Br4923 QNNGQSCIAAKRFIAHADIYDEFVEKFVARMETLRVGDPTDPDTDVGPLA
*: * . .* . : : . . * :**:
Mb2883c|M.bovis_AF2122/97 SRAHRDKVAGYVP----DDAPVAFRGTAPAGRG----FWFPPTVLT-PKR
Rv2858c|M.tuberculosis_H37Rv SRAHRDKVAGYVP----DDAPVAFRGTAPAGRG----FWFPPTVLT-PKR
MAV_1897|M.avium_104 NQPQYEKVLSHFASAREQGATVAYGGEPAGELGG---FFVKPTVLTGVDR
MMAR_3556|M.marinum_M SLAHRDRVADIVAKAPAQGARIVVGGRIPDLAG----AFYLPTLLADVGE
MUL_2789|M.ulcerans_Agy99 SLAHRDRVADILAKAPAQGARIVVGGRIPDLAG----AFYLPTLLADVGE
Mflv_1858|M.gilvum_PYR-GCK NAKQVATVTELVSDAVSRGATVAVGGVAPGG----PGNFYPATVLTDVPA
Mvan_4876|M.vanbaalenii_PYR-1 NAKQVATVTELVSDAVSRGATVAVGGVAPGG----PGNFYPATVLTDVPA
MAB_4208|M.abscessus_ATCC_1997 TAKQRASVDELVQDAVSKGAVVTTGGVVADG----EGYFYPPTVLTDVPA
MSMEG_3007|M.smegmatis_MC2_155 SARQLDGVSELVDTLRGLDSELLTGGNALPG----DGFFFAPTVFRVNGR
TH_4137|M.thermoresistible__bu DEVALKKMQRQVDDAVDKGAKLELGGSRLQGPAFDNGHFYAPTILTGVTP
MLBr_02573|M.leprae_Br4923 TESGRAQVEQQVDAAAAAGAVIRCGGKRPDR----PGWFYPPTVITDITK
: . .: : * : .*::
Mb2883c|M.bovis_AF2122/97 GDRTVTDEIFGPVVVVLTFDDEADAISLANDTAYGLSGSIWTDDLSRALR
Rv2858c|M.tuberculosis_H37Rv GDRTVTDEIFGPVVVVLTFDDEADAISLANDTAYGLSGSIWTDDLSRALR
MAV_1897|M.avium_104 SMRAVAEEVFGPVLAVMTFTDEDEAIAAANDTEFGLAAGVWTKDVHRAHR
MMAR_3556|M.marinum_M QSQAYREEIFGPVLTVRAHDGDDDALRQANDTDYGLAASAWTRDVYRAQR
MUL_2789|M.ulcerans_Agy99 QSQAYREEIFGPVLTVRAHDGDDDALRQANDTDYGLAASAWTRDVYRAQR
Mflv_1858|M.gilvum_PYR-GCK DARILKEEVFGPVAPIAGFDTEEEGIAAANDTEYGLAAYVYTQSLDRALR
Mvan_4876|M.vanbaalenii_PYR-1 DARILKEEVFGPVAPIAGFDTEEEGIAAANDTEYGLAAYVYTQSLDRALR
MAB_4208|M.abscessus_ATCC_1997 DARILREEVFGPVAAITGFDGEEAGIAAANDTEFGLAAYVYTANLDRALR
MSMEG_3007|M.smegmatis_MC2_155 PTELCSRELFAPVATIYQVESTSEAIEFANDTRYGLAAYVFTRDLSRAVA
TH_4137|M.thermoresistible__bu DMLICSEETFGPIAPVMVFDDEDEAIAMANSTEYGLAGYIYTRDISRAIR
MLBr_02573|M.leprae_Br4923 DMALYTDEVFGPVASVYCATNIDDAIEIANATPFGLGSNAWTQNESEQRH
* *.*: : .: ** * :**.. :* . .
Mb2883c|M.bovis_AF2122/97 VARAVESGNLSVNSHSSVRFNTPFGGFKQSGVGRELGPDAPLQFTETKNV
Rv2858c|M.tuberculosis_H37Rv VARAVESGNLSVNSHSSVRFNTPFGGFKQSGVGRELGPDAPLQFTETKNV
MAV_1897|M.avium_104 VAARLNAGTVWVNAYRVVSPHVPFGGMGHSGIGRENGIDAVKDFTETKAV
MMAR_3556|M.marinum_M ASREISAGCVWINDHIPIVSEMPHGGVAASGFGKDMSEYSFEEYLTIKHV
MUL_2789|M.ulcerans_Agy99 ASREISAGCVWINDHIPIVSEMPHGGVAASGFGNDMSEYSFDEYLTIKHV
Mflv_1858|M.gilvum_PYR-GCK VAEGIESGMVGVNRGVISDPAAPFGGIKESGFGREGGTEGIEEYLDTKYI
Mvan_4876|M.vanbaalenii_PYR-1 VAENIESGMVGVNRGVISDAAAPFGGIKESGFGREGGTEGIEEYLDTKYI
MAB_4208|M.abscessus_ATCC_1997 VSQAVESGMVGVNRGVISDVAAPFGGIKESGIGREAGSEGIEEYLETKYI
MSMEG_3007|M.smegmatis_MC2_155 VGERLDFGMVGINRGIMADPAAAFGGVKASGLGREGGHDAIYDYLEPKYL
TH_4137|M.thermoresistible__bu VGEALDFGMIGINDINPTSAAVPFGGIKQSGLGREGARAGIEEYLDTKVL
MLBr_02573|M.leprae_Br4923 FIDNIVAGQVFINGMTVSYPELPFGGIKRSGYGRELSTHGIREFCNIKTV
: * : :* ..**. ** *.: . . :: * :
Mb2883c|M.bovis_AF2122/97 FIAVGEEM-------------
Rv2858c|M.tuberculosis_H37Rv FIAVGEEM-------------
MAV_1897|M.avium_104 WVELSGATRDPFTLG------
MMAR_3556|M.marinum_M MSDITGVAEKPWHRTIFSTRI
MUL_2789|M.ulcerans_Agy99 MSDITGVAEKPWHRTIFSTRI
Mflv_1858|M.gilvum_PYR-GCK ALTK-----------------
Mvan_4876|M.vanbaalenii_PYR-1 ALTR-----------------
MAB_4208|M.abscessus_ATCC_1997 ALP------------------
MSMEG_3007|M.smegmatis_MC2_155 ALTVDEEAGLA----------
TH_4137|M.thermoresistible__bu GIAL-----------------
MLBr_02573|M.leprae_Br4923 WIA------------------