For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MDTTPTRELFDEAYESRTAPWVIGEPQPAVVELERAGLIRSRVLDVGCGAGEHTILLTRLGYDVLGIDFS PQAIEMARENARGRGVDARFAVGDAMALGDLGDGAYDTILDSALFHIFDDADRQTYVASLHAGCRPGGTV HILALSDAGRGFGPEVSEEQIRKAFGDGWDLEALETTTYRGVVGPVHAEAIGLPVGTQVDEPAWLARARR L
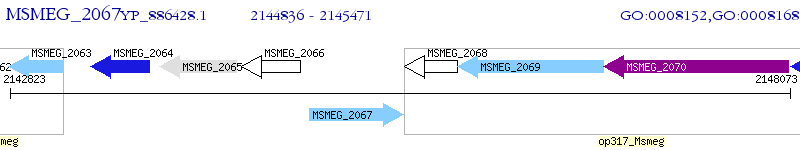
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2067 | - | - | 100% (211) | methyltransferase type 12 |
| M. smegmatis MC2 155 | MSMEG_3041 | - | 5e-28 | 39.88% (168) | thiopurine S-methyltransferase (tpmt) superfamily protein |
| M. smegmatis MC2 155 | MSMEG_6235 | - | 2e-24 | 35.18% (199) | thiopurine S-methyltransferase (tpmt) superfamily protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1412c | - | 6e-32 | 37.81% (201) | putative transferase |
| M. gilvum PYR-GCK | Mflv_4471 | - | 7e-79 | 69.16% (214) | methyltransferase type 11 |
| M. tuberculosis H37Rv | Rv1377c | - | 7e-32 | 37.81% (201) | putative transferase |
| M. leprae Br4923 | MLBr_00551 | - | 1e-08 | 27.78% (108) | putative methyltransferase |
| M. abscessus ATCC 19977 | MAB_0868c | - | 3e-28 | 34.90% (192) | hypothetical protein MAB_0868c |
| M. marinum M | MMAR_2193 | - | 2e-27 | 34.85% (198) | NodS-like (sam)-dependent methyltransferase |
| M. avium 104 | MAV_3570 | - | 2e-24 | 36.22% (185) | thiopurine S-methyltransferase (tpmt) superfamily protein |
| M. thermoresistible (build 8) | TH_1722 | - | 7e-26 | 39.88% (163) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3309 | - | 2e-24 | 36.92% (195) | methyltransferase |
| M. vanbaalenii PYR-1 | Mvan_1888 | - | 4e-81 | 69.67% (211) | methyltransferase type 11 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1412c|M.bovis_AF2122/97 -----------MPGIDFDALYRGESPGEGLPPITTPPWDTKAPKDNVIGW
Rv1377c|M.tuberculosis_H37Rv -----------MPGIDFDALYRGESPGEGLPPITTPPWDTKAPKDNVIGW
MMAR_2193|M.marinum_M --------------MDFDALYRGESPRPGVAPMTTPPWDTKAPKENVIAW
MAV_3570|M.avium_104 -MTASFDPADPAR---FEEMYRDQRTSHGLPAA--TPWDIGGPQPVVRQL
MUL_3309|M.ulcerans_Agy99 -MTAP-DPTDPTR---FEEMYRDQRVAHGLPAA--TPWDIGGPQPVVEQL
MAB_0868c|M.abscessus_ATCC_199 -MTASTQPPDVPSQETFEAVYNGKPMFEGAPAAG-VPWDIRDAQPRIKEL
Mflv_4471|M.gilvum_PYR-GCK ---------MDMDQTPTLSRFDDFYKDE----DKTPPWVIGEPQPAVVDI
Mvan_1888|M.vanbaalenii_PYR-1 -----------MDLTPRLSRFDEFYKN------QTPPWVIGEPQQAIVEL
MSMEG_2067|M.smegmatis_MC2_155 -----------MDTTPTRELFDEAYES------RTAPWVIGEPQPAVVEL
TH_1722|M.thermoresistible__bu MATEDPGTGDPTDTIDWDSVYRQEAPGF----DGPPPWNIGEPQPELAAL
MLBr_00551|M.leprae_Br4923 -------MNLYTPIHEDQSLAAIHRAMWALGDYALMAEEVMAPLGPILVT
. . :
Mb1412c|M.bovis_AF2122/97 HTGGWVHGDVLDIGCGLGDNAIYLARNGYQVTGLDISPTALTTAKRRASD
Rv1377c|M.tuberculosis_H37Rv HTGGWVHGDVLDIGCGLGDNAIYLARNGYQVTGLDISPTALTTAKRRASD
MMAR_2193|M.marinum_M QAGGWVHGDVLDIGCGLGDNAIYLAAHGHPVTGLDISPTALITAARRADD
MAV_3570|M.avium_104 VALGAVKGEVLDPGTGPGHHAIYYASQGFSATGIDGSAAAIERARANART
MUL_3309|M.ulcerans_Agy99 VALGAVKGEVLDPGTGPGHHAIYYASKGYSATGIDGSATAIERARDNAAK
MAB_0868c|M.abscessus_ATCC_199 AAYGALRGAILDIGCGLGENSIYLTQQGLSVTGLDFSPSAIEQAKQRAAA
Mflv_4471|M.gilvum_PYR-GCK ERAGLITGRVLDVGCGTGERTILLTAAGYDVLGVDGAPSAVEQARHNAKT
Mvan_1888|M.vanbaalenii_PYR-1 EQAGLIGGRVLDVGCGTGEHTILLARAGYDVLGIDGAPTAVEQARRNAEA
MSMEG_2067|M.smegmatis_MC2_155 ERAGLIRSRVLDVGCGAGEHTILLTRLGYDVLGIDFSPQAIEMARENARG
TH_1722|M.thermoresistible__bu HRAGAFRSDVLDAGCGHAEISMALAADGYTVVGIDLSPTAIAAANRTAQE
MLBr_00551|M.leprae_Br4923 ATGIRPGMQVLDVAAGSGNISLPAAKTGATIVSTDLTPELLQRSQARATE
:** . * .. :: : * . * :. : : *
Mb1412c|M.bovis_AF2122/97 AGVDVK-FAVGDATKLTG-YTGAFDTVIDCGMFHCLDDDG--KRSYAASV
Rv1377c|M.tuberculosis_H37Rv AGVDVK-FAVGDATKLTG-YTGAFDTVIDCGMFHCLDDDG--KRSYAASV
MMAR_2193|M.marinum_M AGVHVT-FAVADATKLDG-YTDAFDTVIDSGMFHCLDDEG--KRSYAAAV
MAV_3570|M.avium_104 AGVSVN-FELADATKLDG-FDGRFDTVVDCAFYHVFATEPELRSSYARAL
MUL_3309|M.ulcerans_Agy99 AGVSVN-FQVADATRLEG-LDNRFDTVVDCAFYHTFGTAPELQRSYVQAL
MAB_0868c|M.abscessus_ATCC_199 AGVTVD-LHVADATKLDG-WDAQFDTVIDSAVYHCFDHEG--HRQYASAL
Mflv_4471|M.gilvum_PYR-GCK RVVDAR-FDVADALDLGD--EPAYDTIIDSALFHIFDDAD--REKYVRSL
Mvan_1888|M.vanbaalenii_PYR-1 QGVDAR-FELADALHLGP--DPTYDTIVDSALFHIFDDAD--RATYVRSL
MSMEG_2067|M.smegmatis_MC2_155 RGVDAR-FAVGDAMALGDLGDGAYDTILDSALFHIFDDAD--RQTYVASL
TH_1722|M.thermoresistible__bu RGLATASFVQGDITDFGG-YDGRFNTIIDSTLFHSIPVQA--RDGYLRSI
MLBr_00551|M.leprae_Br4923 LGLTLE-YREADAQALPF-GDGEFDVVMSAIGVMFAPDHR----RAASEL
: .* : ::.::.. :
Mb1412c|M.bovis_AF2122/97 HRATRPGATLLLSCFSNA------MPPDEEWPRSTVSEQTLRDVLGGAGW
Rv1377c|M.tuberculosis_H37Rv HRATRPGATLLLSCFSNA------MPPDEEWPRSTVSEQTLRDVLGGAGW
MMAR_2193|M.marinum_M HRATRPGATLLMSCFCDA------NPPNEQQPRPAVPEQTLRDVLGGAGW
MAV_3570|M.avium_104 HRATKPGARLYMFEFGEH------DVNGFKMMRSLSE-NDFRDVLPAAGW
MUL_3309|M.ulcerans_Agy99 WRATKPGARLYMYEFGAH------QVNGLTAIRSLPE-ENFREMLPLGGW
MAB_0868c|M.abscessus_ATCC_199 HRATRPGARWFIACFGDG------SVNGITMPFGVQDPDEIKSVLSEARW
Mflv_4471|M.gilvum_PYR-GCK GRATAPGSVVHLLALSD---------TGRGFGP-EVSEETIRAAFADPGW
Mvan_1888|M.vanbaalenii_PYR-1 HAATRPGSVVHLLALSD---------SGRGFGP-EVSEHTIRAAFG-AGW
MSMEG_2067|M.smegmatis_MC2_155 HAGCRPGGTVHILALSD---------AGRGFGP-EVSEEQIRKAFG-DGW
TH_1722|M.thermoresistible__bu HRAAAPGARYFALVFAKGAFP---EEMEQGPGPNPVDEDELREAVS-KYW
MLBr_00551|M.leprae_Br4923 VRVCRPNGTIGVISWTQEGFFGRMLATIRPYRPSLSAALPPAALWGREAY
*.. :
Mb1412c|M.bovis_AF2122/97 DIESLEPATVRREL------------------------------------
Rv1377c|M.tuberculosis_H37Rv DIESLEPATVRREL------------------------------------
MMAR_2193|M.marinum_M EIAALDPVTVNRQLGGS---------------------------------
MAV_3570|M.avium_104 EISYLGPTTYQVNLSAETIQMMAARNPD----------------------
MUL_3309|M.ulcerans_Agy99 EITYLGPTTYRTTVSMETLETMAARNPE----------------------
MAB_0868c|M.abscessus_ATCC_199 NIQYFGRTTYLGSRAAFVGAALAGIDPEQMPADMRERY---------QRP
Mflv_4471|M.gilvum_PYR-GCK DIEALTTTYYRG--VITAPHADAMGLDV----------------------
Mvan_1888|M.vanbaalenii_PYR-1 EVEALTETTYRG--VVIDAHTEALNLPA----------------------
MSMEG_2067|M.smegmatis_MC2_155 DLEALETTTYRG--VVGPVHAEAIGLPV----------------------
TH_1722|M.thermoresistible__bu EIDEIRPAFIHANMPPDVPGVESFDTDE----------------------
MLBr_00551|M.leprae_Br4923 IRGLLDDTVTNVKTERRQLEVKRFNSAEGVHNYFKNHYGPTIEAYTNIGD
: .
Mb1412c|M.bovis_AF2122/97 --------------------DGTEVEMAFWNVRAQRRGS
Rv1377c|M.tuberculosis_H37Rv --------------------DGTEVEMAFWNVRAQRRGS
MMAR_2193|M.marinum_M --------------------DGQPIKLAFWYVRARRQN-
MAV_3570|M.avium_104 -MADEAAKLLERFRAMEPWLQAGRVHAPFWEVHATRVD-
MUL_3309|M.ulcerans_Agy99 -VADQVGPLVERFKTIQPWLTDGIAHVPFWEVHATRPD-
MAB_0868c|M.abscessus_ATCC_199 GVAEQAQQTFERLQQLD----DDMIHMPFFNIVAERG--
Mflv_4471|M.gilvum_PYR-GCK ---------------------GTVVDEPAWLARIRRL--
Mvan_1888|M.vanbaalenii_PYR-1 ---------------------GTVVDEPAWSARIRRL--
MSMEG_2067|M.smegmatis_MC2_155 ---------------------GTQVDEPAWLARARRL--
TH_1722|M.thermoresistible__bu ---------------------KGRTKLPAFLLSAHKA--
MLBr_00551|M.leprae_Br4923 NSVLAAELDAQLTELAQKYLTNSTMGWEYLLLTAQKH--
: