For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DMDQTPTLSRFDDFYKDEDKTPPWVIGEPQPAVVDIERAGLITGRVLDVGCGTGERTILLTAAGYDVLGV DGAPSAVEQARHNAKTRVVDARFDVADALDLGDEPAYDTIIDSALFHIFDDADREKYVRSLGRATAPGSV VHLLALSDTGRGFGPEVSEETIRAAFADPGWDIEALTTTYYRGVITAPHADAMGLDVGTVVDEPAWLARI RRL*
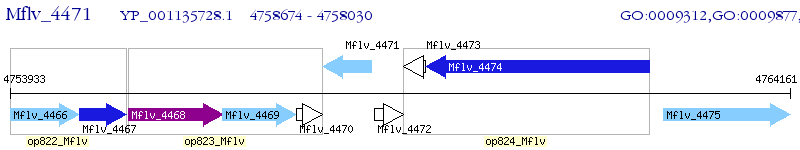
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4471 | - | - | 100% (214) | methyltransferase type 11 |
| M. gilvum PYR-GCK | Mflv_1324 | - | 3e-20 | 37.50% (176) | methyltransferase type 11 |
| M. gilvum PYR-GCK | Mflv_3090 | - | 1e-09 | 38.21% (123) | methyltransferase type 11 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1412c | - | 2e-27 | 34.86% (218) | putative transferase |
| M. tuberculosis H37Rv | Rv1377c | - | 2e-27 | 34.86% (218) | putative transferase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0961c | - | 3e-27 | 42.07% (164) | hypothetical protein MAB_0961c |
| M. marinum M | MMAR_2041 | - | 1e-26 | 34.30% (207) | methyltransferase |
| M. avium 104 | MAV_3570 | - | 9e-28 | 33.18% (211) | thiopurine S-methyltransferase (tpmt) superfamily protein |
| M. smegmatis MC2 155 | MSMEG_2067 | - | 8e-79 | 69.16% (214) | methyltransferase type 12 |
| M. thermoresistible (build 8) | TH_1722 | - | 1e-24 | 36.65% (191) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3309 | - | 2e-26 | 33.82% (207) | methyltransferase |
| M. vanbaalenii PYR-1 | Mvan_1888 | - | 3e-87 | 74.06% (212) | methyltransferase type 11 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1412c|M.bovis_AF2122/97 -----------MPGIDFDALYRGESPGEGLPPITTPPWDTKAPKDNVIGW
Rv1377c|M.tuberculosis_H37Rv -----------MPGIDFDALYRGESPGEGLPPITTPPWDTKAPKDNVIGW
MMAR_2041|M.marinum_M ----MTAP-DPTDPTRFEEMYRDQRVAHGLPAAT--PWDIGGPQPVVEQL
MUL_3309|M.ulcerans_Agy99 ----MTAP-DPTDPTRFEEMYRDQRVAHGLPAAT--PWDIGGPQPVVEQL
MAV_3570|M.avium_104 ----MTASFDPADPARFEEMYRDQRTSHGLPAAT--PWDIGGPQPVVRQL
Mflv_4471|M.gilvum_PYR-GCK -----MDMDQTPTLSRFDDFYKDED--------KTPPWVIGEPQPAVVDI
Mvan_1888|M.vanbaalenii_PYR-1 -------MDLTPRLSRFDEFYKN----------QTPPWVIGEPQQAIVEL
MSMEG_2067|M.smegmatis_MC2_155 -------MDTTPTRELFDEAYES----------RTAPWVIGEPQPAVVEL
MAB_0961c|M.abscessus_ATCC_199 MTSN--------ELPDWDATYQGKGELFQ----GEPPWNIGEPQPELAAL
TH_1722|M.thermoresistible__bu MATEDPGTGDPTDTIDWDSVYRQEAPGFD----GPPPWNIGEPQPELAAL
:: *. ** *: :
Mb1412c|M.bovis_AF2122/97 HTGGWVHGDVLDIGCGLGDNAIYLARNGYQVTGLDISPTALTTAKRRASD
Rv1377c|M.tuberculosis_H37Rv HTGGWVHGDVLDIGCGLGDNAIYLARNGYQVTGLDISPTALTTAKRRASD
MMAR_2041|M.marinum_M VALGAVKGEVLDPGTGPGHHAIYYASKGYSATGIDGSATAIERARDNAAK
MUL_3309|M.ulcerans_Agy99 VALGAVKGEVLDPGTGPGHHAIYYASKGYSATGIDGSATAIERARDNAAK
MAV_3570|M.avium_104 VALGAVKGEVLDPGTGPGHHAIYYASQGFSATGIDGSAAAIERARANART
Mflv_4471|M.gilvum_PYR-GCK ERAGLITGRVLDVGCGTGERTILLTAAGYDVLGVDGAPSAVEQARHNAKT
Mvan_1888|M.vanbaalenii_PYR-1 EQAGLIGGRVLDVGCGTGEHTILLARAGYDVLGIDGAPTAVEQARRNAEA
MSMEG_2067|M.smegmatis_MC2_155 ERAGLIRSRVLDVGCGAGEHTILLTRLGYDVLGIDFSPQAIEMARENARG
MAB_0961c|M.abscessus_ATCC_199 IDAGKFHGTVLDVGCGHAETSLRLAALGHTTVGLDLSPTAIEAARAAAAE
TH_1722|M.thermoresistible__bu HRAGAFRSDVLDAGCGHAEISMALAADGYTVVGIDLSPTAIAAANRTAQE
* . . *** * * .. :: : *. . *:* :. *: *. *
Mb1412c|M.bovis_AF2122/97 AGVD-VKFAVGDATKLTG-YTGAFDTVIDCGMFHCLDDD--GKRSYAASV
Rv1377c|M.tuberculosis_H37Rv AGVD-VKFAVGDATKLTG-YTGAFDTVIDCGMFHCLDDD--GKRSYAASV
MMAR_2041|M.marinum_M AGVS-VNFQVADATRLEG-LDNRFDTVVDCAFYHTFGTAPELQRSYVRAL
MUL_3309|M.ulcerans_Agy99 AGVS-VNFQVADATRLEG-LDNRFDTVVDCAFYHTFGTAPELQRSYVQAL
MAV_3570|M.avium_104 AGVS-VNFELADATKLDG-FDGRFDTVVDCAFYHVFATEPELRSSYARAL
Mflv_4471|M.gilvum_PYR-GCK RVVD-ARFDVADALDLGD--EPAYDTIIDSALFHIFDDAD--REKYVRSL
Mvan_1888|M.vanbaalenii_PYR-1 QGVD-ARFELADALHLGP--DPTYDTIVDSALFHIFDDAD--RATYVRSL
MSMEG_2067|M.smegmatis_MC2_155 RGVD-ARFAVGDAMALGDLGDGAYDTILDSALFHIFDDAD--RQTYVASL
MAB_0961c|M.abscessus_ATCC_199 RGLTNATFEVADITDFSG-YDGRFNTIVDSTLFHSIPVEA--REAYLQSI
TH_1722|M.thermoresistible__bu RGLATASFVQGDITDFGG-YDGRFNTIIDSTLFHSIPVQA--RDGYLRSI
: . * .* : ::*::*. ::* : : * ::
Mb1412c|M.bovis_AF2122/97 HRATRPGATLLLSCFSNAMPPDE--EWPRSTVSEQTLRDVLGGAGWDIES
Rv1377c|M.tuberculosis_H37Rv HRATRPGATLLLSCFSNAMPPDE--EWPRSTVSEQTLRDVLGGAGWDIES
MMAR_2041|M.marinum_M WRATKPGARLYMYEFGAHQVNGL--TAIRS-LPEENFREMLPLGGWEITY
MUL_3309|M.ulcerans_Agy99 WRATKPGARLYMYEFGAHQVNGL--TAIRS-LPEENFREMLPLGGWEITY
MAV_3570|M.avium_104 HRATKPGARLYMFEFGEHDVNGF--KMMRS-LSENDFRDVLPAAGWEISY
Mflv_4471|M.gilvum_PYR-GCK GRATAPGSVVHLLALSDTG------RGFGPEVSEETIRAAFADPGWDIEA
Mvan_1888|M.vanbaalenii_PYR-1 HAATRPGSVVHLLALSDSG------RGFGPEVSEHTIRAAFG-AGWEVEA
MSMEG_2067|M.smegmatis_MC2_155 HAGCRPGGTVHILALSDAG------RGFGPEVSEEQIRKAFG-DGWDLEA
MAB_0961c|M.abscessus_ATCC_199 SRAAAPDASYFVLVFAVGAFP----PGIGPNAVTEDELRAAVEPYWAIDE
TH_1722|M.thermoresistible__bu HRAAAPGARYFALVFAKGAFPEEMEQGPGPNPVDEDELREAVSKYWEIDE
. *.. :. . . * :
Mb1412c|M.bovis_AF2122/97 LEPATVRRELDGTEVEMA--------------------------------
Rv1377c|M.tuberculosis_H37Rv LEPATVRRELDGTEVEMA--------------------------------
MMAR_2041|M.marinum_M LGPTTYRTTVSMETLETMAARNPEVADQVGPLVERFKTIQPWLTDGIAHV
MUL_3309|M.ulcerans_Agy99 LGPTTYRTTVSMETLETMAARNPEVADQVGPLVERFKTIQPWLTDGIAHV
MAV_3570|M.avium_104 LGPTTYQVNLSAETIQMMAARNPDMADEAAKLLERFRAMEPWLQAGRVHA
Mflv_4471|M.gilvum_PYR-GCK LTTTYYRGVITAPHADAMG----------------------LDVGTVVDE
Mvan_1888|M.vanbaalenii_PYR-1 LTETTYRGVVIDAHTEALN----------------------LPAGTVVDE
MSMEG_2067|M.smegmatis_MC2_155 LETTTYRGVVGPVHAEAIG----------------------LPVGTQVDE
MAB_0961c|M.abscessus_ATCC_199 LSSAYIHSNIPEVIPGANFE----------------MPVFHKDEKGRNKQ
TH_1722|M.thermoresistible__bu IRPAFIHANMPPDVPG--------------------VESFDTDEKGRTKL
: : : :
Mb1412c|M.bovis_AF2122/97 -FWNVRAQRRGS
Rv1377c|M.tuberculosis_H37Rv -FWNVRAQRRGS
MMAR_2041|M.marinum_M PFWEVHATRLD-
MUL_3309|M.ulcerans_Agy99 PFWEVHATRPD-
MAV_3570|M.avium_104 PFWEVHATRVD-
Mflv_4471|M.gilvum_PYR-GCK PAWLARIRRL--
Mvan_1888|M.vanbaalenii_PYR-1 PAWSARIRRL--
MSMEG_2067|M.smegmatis_MC2_155 PAWLARARRL--
MAB_0961c|M.abscessus_ATCC_199 PAFLLQAHKR--
TH_1722|M.thermoresistible__bu PAFLLSAHKA--
: :