For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TASTQPPDVPSQETFEAVYNGKPMFEGAPAAGVPWDIRDAQPRIKELAAYGALRGAILDIGCGLGENSIY LTQQGLSVTGLDFSPSAIEQAKQRAAAAGVTVDLHVADATKLDGWDAQFDTVIDSAVYHCFDHEGHRQYA SALHRATRPGARWFIACFGDGSVNGITMPFGVQDPDEIKSVLSEARWNIQYFGRTTYLGSRAAFVGAALA GIDPEQMPADMRERYQRPGVAEQAQQTFERLQQLDDDMIHMPFFNIVAERG*
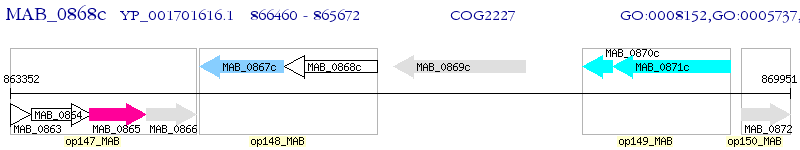
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0868c | - | - | 100% (262) | hypothetical protein MAB_0868c |
| M. abscessus ATCC 19977 | MAB_0867c | - | 7e-70 | 53.36% (253) | putative transferase |
| M. abscessus ATCC 19977 | MAB_0240 | - | 3e-29 | 40.00% (190) | hypothetical protein MAB_0240 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2694c | - | 2e-42 | 39.70% (267) | hypothetical protein Mb2694c |
| M. gilvum PYR-GCK | Mflv_1324 | - | 2e-26 | 36.69% (169) | methyltransferase type 11 |
| M. tuberculosis H37Rv | Rv2675c | - | 2e-42 | 39.70% (267) | hypothetical protein Rv2675c |
| M. leprae Br4923 | MLBr_00551 | - | 8e-09 | 33.33% (93) | putative methyltransferase |
| M. marinum M | MMAR_2041 | - | 8e-42 | 38.85% (260) | methyltransferase |
| M. avium 104 | MAV_3570 | - | 1e-45 | 40.82% (267) | thiopurine S-methyltransferase (tpmt) superfamily protein |
| M. smegmatis MC2 155 | MSMEG_3041 | - | 8e-40 | 41.75% (206) | thiopurine S-methyltransferase (tpmt) superfamily protein |
| M. thermoresistible (build 8) | TH_1722 | - | 1e-23 | 37.33% (150) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3309 | - | 4e-42 | 38.85% (260) | methyltransferase |
| M. vanbaalenii PYR-1 | Mvan_1888 | - | 1e-26 | 37.35% (166) | methyltransferase type 11 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1324|M.gilvum_PYR-GCK MA----------EVMDWDSAYRGEGE-FEGEP----PWNIGEPQPELAAL
TH_1722|M.thermoresistible__bu MATEDPGTGDPTDTIDWDSVYRQEAPGFDGPP----PWNIGEPQPELAAL
Mb2694c|M.bovis_AF2122/97 -MTAQFDPADPTR---FEEMYRDDRVAHGLPAA--TPWDIGGPQPVVQQL
Rv2675c|M.tuberculosis_H37Rv -MTAQFDPADPTR---FEEMYRDDRVAHGLPAA--TPWDIGGPQPVVQQL
MAV_3570|M.avium_104 -MTASFDPADPAR---FEEMYRDQRTSHGLPAA--TPWDIGGPQPVVRQL
MMAR_2041|M.marinum_M -MTAP-DPTDPTR---FEEMYRDQRVAHGLPAA--TPWDIGGPQPVVEQL
MUL_3309|M.ulcerans_Agy99 -MTAP-DPTDPTR---FEEMYRDQRVAHGLPAA--TPWDIGGPQPVVEQL
MAB_0868c|M.abscessus_ATCC_199 -MTASTQPPDVPSQETFEAVYNGKPMFEGAPAAG-VPWDIRDAQPRIKEL
MSMEG_3041|M.smegmatis_MC2_155 -MTGP--TGSEFD---FDALYRGESPAEGVPPITSVPWDTKAPKENVVAW
Mvan_1888|M.vanbaalenii_PYR-1 ----MDLTPRLSR---FDEFYKNQT----------PPWVIGEPQQAIVEL
MLBr_00551|M.leprae_Br4923 -MNLYTPIHEDQS---LAAIHRAMWALGDYALM---AEEVMAPLGPILVT
:. . . :
Mflv_1324|M.gilvum_PYR-GCK WRDGRFRSEVLDAGCGHAELSLALAADGYTVTGVDISPTAMAAASAAAAE
TH_1722|M.thermoresistible__bu HRAGAFRSDVLDAGCGHAEISMALAADGYTVVGIDLSPTAIAAANRTAQE
Mb2694c|M.bovis_AF2122/97 VALGAIRGEVLDPGTGPGHHAIYYAAKGYAATGIDGSVAAIERARDNARK
Rv2675c|M.tuberculosis_H37Rv VALGAIRGEVLDPGTGPGHHAIYYAAKGYAATGIDGSVAAIERARDNARK
MAV_3570|M.avium_104 VALGAVKGEVLDPGTGPGHHAIYYASQGFSATGIDGSAAAIERARANART
MMAR_2041|M.marinum_M VALGAVKGEVLDPGTGPGHHAIYYASKGYSATGIDGSATAIERARDNAAK
MUL_3309|M.ulcerans_Agy99 VALGAVKGEVLDPGTGPGHHAIYYASKGYSATGIDGSATAIERARDNAAK
MAB_0868c|M.abscessus_ATCC_199 AAYGALRGAILDIGCGLGENSIYLTQQGLSVTGLDFSPSAIEQAKQRAAA
MSMEG_3041|M.smegmatis_MC2_155 EQAGLVHGHVLDIGCGLGDNAIYLAQQGYRVTALDISPTALITAERRAAD
Mvan_1888|M.vanbaalenii_PYR-1 EQAGLIGGRVLDVGCGTGEHTILLARAGYDVLGIDGAPTAVEQARRNAEA
MLBr_00551|M.leprae_Br4923 ATGIRPGMQVLDVAAGSGNISLPAAKTGATIVSTDLTPELLQRSQARATE
:** . * .. :: : * . * : : : *
Mflv_1324|M.gilvum_PYR-GCK RGLSEKASFVAADITALSGFDGRFSTVVDST-------------------
TH_1722|M.thermoresistible__bu RGLAT-ASFVQGDITDFGGYDGRFNTIIDST-------------------
Mb2694c|M.bovis_AF2122/97 AGVSV--NFQVGDATTLDGLDGRFDTVVDCA-------------------
Rv2675c|M.tuberculosis_H37Rv AGVSV--NFQVGDATTLDGLDGRFDTVVDCA-------------------
MAV_3570|M.avium_104 AGVSV--NFELADATKLDGFDGRFDTVVDCA-------------------
MMAR_2041|M.marinum_M AGVSV--NFQVADATRLEGLDNRFDTVVDCA-------------------
MUL_3309|M.ulcerans_Agy99 AGVSV--NFQVADATRLEGLDNRFDTVVDCA-------------------
MAB_0868c|M.abscessus_ATCC_199 AGVTV--DLHVADATKLDGWDAQFDTVIDSA-------------------
MSMEG_3041|M.smegmatis_MC2_155 AGVDV--TFAVADATRLDGYDDVFDTVVDSG-------------------
Mvan_1888|M.vanbaalenii_PYR-1 QGVDA--RFELADALHLG-PDPTYDTIVDSA-------------------
MLBr_00551|M.leprae_Br4923 LGLTL--EYREADAQALPFGDGEFDVVMSAIGVMFAPDHRRAASELVRVC
*: .* : * :..::..
Mflv_1324|M.gilvum_PYR-GCK ----------------LFHSLP--VESRAGYLESVHRAARPGAGFFVLVF
TH_1722|M.thermoresistible__bu ----------------LFHSIP--VQARDGYLRSIHRAAAPGARYFALVF
Mb2694c|M.bovis_AF2122/97 ----------------FYHTFSTAPELQRCYVRALRRASKPGARLYMFEF
Rv2675c|M.tuberculosis_H37Rv ----------------FYHTFSTAPELQRCYVRALRRASKPGARLYMFEF
MAV_3570|M.avium_104 ----------------FYHVFATEPELRSSYARALHRATKPGARLYMFEF
MMAR_2041|M.marinum_M ----------------FYHTFGTAPELQRSYVRALWRATKPGARLYMYEF
MUL_3309|M.ulcerans_Agy99 ----------------FYHTFGTAPELQRSYVQALWRATKPGARLYMYEF
MAB_0868c|M.abscessus_ATCC_199 ----------------VYHCFD--HEGHRQYASALHRATRPGARWFIACF
MSMEG_3041|M.smegmatis_MC2_155 ----------------MFHCLS--DDAKGSYAAAVHRATRAGATLLLGAF
Mvan_1888|M.vanbaalenii_PYR-1 ----------------LFHIFD--DADRATYVRSLHAATRPGSVVHLLAL
MLBr_00551|M.leprae_Br4923 RPNGTIGVISWTQEGFFGRMLATIRPYRPSLSAALPPAALWGREAYIRGL
. : : : :: *: * :
Mflv_1324|M.gilvum_PYR-GCK AKGAFPADTEQGP--NEVSEIELREAVS-PYWTIDDIRPALIHTNVPKLP
TH_1722|M.thermoresistible__bu AKGAFPEEMEQGPGPNPVDEDELREAVS-KYWEIDEIRPAFIHANMP--P
Mb2694c|M.bovis_AF2122/97 GEHNVNGFSMPRS---LSE-DDFRQVLPVGGWEITYLGTTTYQVNLSVEA
Rv2675c|M.tuberculosis_H37Rv GEHNVNGFSMPRS---LSE-DDFRQVLPVGGWEITYLGTTTYQVNLSVEA
MAV_3570|M.avium_104 GEHDVNGFKMMRS---LSE-NDFRDVLPAAGWEISYLGPTTYQVNLSAET
MMAR_2041|M.marinum_M GAHQVNGLTAIRS---LPE-ENFREMLPLGGWEITYLGPTTYRTTVSMET
MUL_3309|M.ulcerans_Agy99 GAHQVNGLTAIRS---LPE-ENFREMLPLGGWEITYLGPTTYRTTVSMET
MAB_0868c|M.abscessus_ATCC_199 GDGSVNGITMPFG---VQDPDEIKSVLSEARWNIQYFGRTTYLGSRAAFV
MSMEG_3041|M.smegmatis_MC2_155 SDANLQHDNWQAA---WVSEDTLRSSLDDGGWDITSLTTETIRVPG----
Mvan_1888|M.vanbaalenii_PYR-1 SDSGRG----FGP---EVSEHTIRAAFG-AGWEVEALTETTYRGVVIDAH
MLBr_00551|M.leprae_Br4923 LDDTVTNVKTERR---QLEVKRFNSAEGVHNYFKNHYGPTIEAYTNIGDN
. :. :
Mflv_1324|M.gilvum_PYR-GCK GMPPPPLDMLD--------------------------------DRGHLRM
TH_1722|M.thermoresistible__bu DVPGVESFDTD--------------------------------EKGRTKL
Mb2694c|M.bovis_AF2122/97 LELMAARNPD--------------MADQVRCVLERFRAIKPWLVGGRVHA
Rv2675c|M.tuberculosis_H37Rv LELMAARNPD--------------MADQVRCVLERFRAIKPWLVGGRVHA
MAV_3570|M.avium_104 IQMMAARNPD--------------MADEAAKLLERFRAMEPWLQAGRVHA
MMAR_2041|M.marinum_M LETMAARNPE--------------VADQVGPLVERFKTIQPWLTDGIAHV
MUL_3309|M.ulcerans_Agy99 LETMAARNPE--------------VADQVGPLVERFKTIQPWLTDGIAHV
MAB_0868c|M.abscessus_ATCC_199 GAALAGIDPEQMPADMRERYQRPGVAEQAQQTFERLQQLD----DDMIHM
MSMEG_3041|M.smegmatis_MC2_155 -AALAG------------------SGDEGG--------------ADEFDA
Mvan_1888|M.vanbaalenii_PYR-1 TEALNLPAGT------------------------------------VVDE
MLBr_00551|M.leprae_Br4923 SVLAAELDAQLT------------------------ELAQKYLTNSTMGW
Mflv_1324|M.gilvum_PYR-GCK QAFLLSAHKEP
TH_1722|M.thermoresistible__bu PAFLLSAHKA-
Mb2694c|M.bovis_AF2122/97 PFWEVHATRVD
Rv2675c|M.tuberculosis_H37Rv PFWEVHATRVD
MAV_3570|M.avium_104 PFWEVHATRVD
MMAR_2041|M.marinum_M PFWEVHATRLD
MUL_3309|M.ulcerans_Agy99 PFWEVHATRPD
MAB_0868c|M.abscessus_ATCC_199 PFFNIVAERG-
MSMEG_3041|M.smegmatis_MC2_155 AFFLVQAQRR-
Mvan_1888|M.vanbaalenii_PYR-1 PAWSARIRRL-
MLBr_00551|M.leprae_Br4923 EYLLLTAQKH-
: