For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DFDALYRGESPRPGVAPMTTPPWDTKAPKENVIAWQAGGWVHGDVLDIGCGLGDNAIYLAAHGHPVTGLD ISPTALITAARRADDAGVHVTFAVADATKLDGYTDAFDTVIDSGMFHCLDDEGKRSYAAAVHRATRPGAT LLMSCFCDANPPNEQQPRPAVPEQTLRDVLGGAGWEIAALDPVTVNRQLGGSDGQPIKLAFWYVRARRQN *
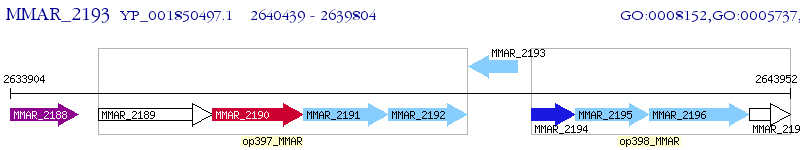
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2193 | - | - | 100% (211) | NodS-like (sam)-dependent methyltransferase |
| M. marinum M | MMAR_2041 | - | 9e-30 | 42.16% (185) | methyltransferase |
| M. marinum M | MMAR_5210 | - | 4e-26 | 35.75% (193) | hypothetical protein MMAR_5210 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1412c | - | 6e-92 | 74.76% (210) | putative transferase |
| M. gilvum PYR-GCK | Mflv_1324 | - | 1e-28 | 36.50% (200) | methyltransferase type 11 |
| M. tuberculosis H37Rv | Rv1377c | - | 6e-92 | 74.76% (210) | putative transferase |
| M. leprae Br4923 | MLBr_00130 | - | 3e-08 | 32.14% (112) | hypothetical protein MLBr_00130 |
| M. abscessus ATCC 19977 | MAB_0868c | - | 1e-39 | 47.16% (176) | hypothetical protein MAB_0868c |
| M. avium 104 | MAV_3570 | - | 8e-32 | 43.24% (185) | thiopurine S-methyltransferase (tpmt) superfamily protein |
| M. smegmatis MC2 155 | MSMEG_3041 | - | 3e-71 | 62.96% (216) | thiopurine S-methyltransferase (tpmt) superfamily protein |
| M. thermoresistible (build 8) | TH_1722 | - | 2e-26 | 33.51% (194) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3309 | - | 6e-30 | 42.16% (185) | methyltransferase |
| M. vanbaalenii PYR-1 | Mvan_1888 | - | 8e-29 | 36.61% (183) | methyltransferase type 11 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1324|M.gilvum_PYR-GCK ---------MA----------EVMDWDS-AYRGEGE-FEGEPPWNIGEPQ
TH_1722|M.thermoresistible__bu ---------MATEDPGTGDPTDTIDWDS-VYRQEAPGFDGPPPWNIGEPQ
Mvan_1888|M.vanbaalenii_PYR-1 -----------------------MDLTPRLSRFDEFYKNQTPPWVIGEPQ
Mb1412c|M.bovis_AF2122/97 ------------MPG-----IDFDALYRGESPGEGLPPITTPPWDTKAPK
Rv1377c|M.tuberculosis_H37Rv ------------MPG-----IDFDALYRGESPGEGLPPITTPPWDTKAPK
MMAR_2193|M.marinum_M --------------------MDFDALYRGESPRPGVAPMTTPPWDTKAPK
MSMEG_3041|M.smegmatis_MC2_155 ------------MTGPTGSEFDFDALYRGESPAEGVPPITSVPWDTKAPK
MAV_3570|M.avium_104 -------MTASFDPADPAR---FEEMYRDQRTSHGLPAA--TPWDIGGPQ
MUL_3309|M.ulcerans_Agy99 -------MTAP-DPTDPTR---FEEMYRDQRVAHGLPAA--TPWDIGGPQ
MAB_0868c|M.abscessus_ATCC_199 -------MTASTQPPDVPSQETFEAVYNGKPMFEGAPAAG-VPWDIRDAQ
MLBr_00130|M.leprae_Br4923 MAFTRIHSFLASAGNTSMYKRVWRFWYPLMTHKLGTDEIMFINWAYEEDP
*
Mflv_1324|M.gilvum_PYR-GCK PELAALWRD------------------GRFRSEVLDAGCGHAELSLALAA
TH_1722|M.thermoresistible__bu PELAALHRA------------------GAFRSDVLDAGCGHAEISMALAA
Mvan_1888|M.vanbaalenii_PYR-1 QAIVELEQA------------------GLIGGRVLDVGCGTGEHTILLAR
Mb1412c|M.bovis_AF2122/97 DNVIGWHTG------------------GWVHGDVLDIGCGLGDNAIYLAR
Rv1377c|M.tuberculosis_H37Rv DNVIGWHTG------------------GWVHGDVLDIGCGLGDNAIYLAR
MMAR_2193|M.marinum_M ENVIAWQAG------------------GWVHGDVLDIGCGLGDNAIYLAA
MSMEG_3041|M.smegmatis_MC2_155 ENVVAWEQA------------------GLVHGHVLDIGCGLGDNAIYLAQ
MAV_3570|M.avium_104 PVVRQLVAL------------------GAVKGEVLDPGTGPGHHAIYYAS
MUL_3309|M.ulcerans_Agy99 PVVEQLVAL------------------GAVKGEVLDPGTGPGHHAIYYAS
MAB_0868c|M.abscessus_ATCC_199 PRIKELAAY------------------GALRGAILDIGCGLGENSIYLTQ
MLBr_00130|M.leprae_Br4923 PMALPLEASDEPNRAHINLYHRTATQVNLSGKRILEVSCGHGGGASYLTR
. :*: . * . : :
Mflv_1324|M.gilvum_PYR-GCK DGY--TVTGVDISPTAMAAASAAAAERGLSEKASFVAADITALSGFDGRF
TH_1722|M.thermoresistible__bu DGY--TVVGIDLSPTAIAAANRTAQERGLAT-ASFVQGDITDFGGYDGRF
Mvan_1888|M.vanbaalenii_PYR-1 AGY--DVLGIDGAPTAVEQARRNAEAQGVDA--RFELADALHLG-PDPTY
Mb1412c|M.bovis_AF2122/97 NGY--QVTGLDISPTALTTAKRRASDAGVDV--KFAVGDATKLTGYTGAF
Rv1377c|M.tuberculosis_H37Rv NGY--QVTGLDISPTALTTAKRRASDAGVDV--KFAVGDATKLTGYTGAF
MMAR_2193|M.marinum_M HGH--PVTGLDISPTALITAARRADDAGVHV--TFAVADATKLDGYTDAF
MSMEG_3041|M.smegmatis_MC2_155 QGY--RVTALDISPTALITAERRAADAGVDV--TFAVADATRLDGYDDVF
MAV_3570|M.avium_104 QGF--SATGIDGSAAAIERARANARTAGVSV--NFELADATKLDGFDGRF
MUL_3309|M.ulcerans_Agy99 KGY--SATGIDGSATAIERARDNAAKAGVSV--NFQVADATRLEGLDNRF
MAB_0868c|M.abscessus_ATCC_199 QGL--SVTGLDFSPSAIEQAKQRAAAAGVTV--DLHVADATKLDGWDAQF
MLBr_00130|M.leprae_Br4923 ALHPASYTGLDLNPAGIKLCQKRHQLPGLEF----VRGDAENLPFDNESF
.:* .:.: . *: .* : :
Mflv_1324|M.gilvum_PYR-GCK STVVDSTLFHSLPVES--RAGYLESVHRAARPGAGFFVLVFAKGAFPADT
TH_1722|M.thermoresistible__bu NTIIDSTLFHSIPVQA--RDGYLRSIHRAAAPGARYFALVFAKGAFPEEM
Mvan_1888|M.vanbaalenii_PYR-1 DTIVDSALFHIFDDAD--RATYVRSLHAATRPGSVVHLLALSD----SGR
Mb1412c|M.bovis_AF2122/97 DTVIDCGMFHCLDDDG--KRSYAASVHRATRPGATLLLSCFSNAMPPDEE
Rv1377c|M.tuberculosis_H37Rv DTVIDCGMFHCLDDDG--KRSYAASVHRATRPGATLLLSCFSNAMPPDEE
MMAR_2193|M.marinum_M DTVIDSGMFHCLDDEG--KRSYAAAVHRATRPGATLLMSCFCDANPPNEQ
MSMEG_3041|M.smegmatis_MC2_155 DTVVDSGMFHCLSDDA--KGSYAAAVHRATRAGATLLLGAFSDANLQHDN
MAV_3570|M.avium_104 DTVVDCAFYHVFATEPELRSSYARALHRATKPGARLYMFEFGEHDVNGFK
MUL_3309|M.ulcerans_Agy99 DTVVDCAFYHTFGTAPELQRSYVQALWRATKPGARLYMYEFGAHQVNGLT
MAB_0868c|M.abscessus_ATCC_199 DTVIDSAVYHCFDHEG--HRQYASALHRATRPGARWFIACFGDGSVNGIT
MLBr_00130|M.leprae_Br4923 DVVINIEASHCYPHFP----RFLAEVVRVLRPGGHLAYADLRPSNKVGEW
..::: * : : . .*. :
Mflv_1324|M.gilvum_PYR-GCK EQGP--NEVSEIELREAVS-PYWTIDDIRPALIHTNVPKL----------
TH_1722|M.thermoresistible__bu EQGPGPNPVDEDELREAVS-KYWEIDEIRPAFIHANMP------------
Mvan_1888|M.vanbaalenii_PYR-1 GFGP---EVSEHTIRAAFG-AGWEVEALTETTYRGVVID-----------
Mb1412c|M.bovis_AF2122/97 WPRS---TVSEQTLRDVLGGAGWDIESLEPATVR----------------
Rv1377c|M.tuberculosis_H37Rv WPRS---TVSEQTLRDVLGGAGWDIESLEPATVR----------------
MMAR_2193|M.marinum_M QPRP---AVPEQTLRDVLGGAGWEIAALDPVTVN----------------
MSMEG_3041|M.smegmatis_MC2_155 WQAA---WVSEDTLRSSLDDGGWDITSLTTETIRV---------------
MAV_3570|M.avium_104 MMRS---LSE-NDFRDVLPAAGWEISYLGPTTYQVNLSAETIQMMAARNP
MUL_3309|M.ulcerans_Agy99 AIRS---LPE-ENFREMLPLGGWEITYLGPTTYRTTVSMETLETMAARNP
MAB_0868c|M.abscessus_ATCC_199 MPFG---VQDPDEIKSVLSEARWNIQYFGRTTYLGSRAAFVGAALAGIDP
MLBr_00130|M.leprae_Br4923 EVDFANSRLQQLSQREINAEVLRGIASNSQKSRDLVDRHLPAFLRFAG--
: :
Mflv_1324|M.gilvum_PYR-GCK ----------------------PGMPPPPLDMLDDRGHLRMQAFLLSAHK
TH_1722|M.thermoresistible__bu ----------------------PDVPGVESFDTDEKGRTKLPAFLLSAHK
Mvan_1888|M.vanbaalenii_PYR-1 -------------------------AHTEALNLPAGTVVDEPAWSARIRR
Mb1412c|M.bovis_AF2122/97 ------------------------REL-------DGTEVEMAFWNVRAQR
Rv1377c|M.tuberculosis_H37Rv ------------------------REL-------DGTEVEMAFWNVRAQR
MMAR_2193|M.marinum_M ------------------------RQLGGS----DGQPIKLAFWYVRARR
MSMEG_3041|M.smegmatis_MC2_155 ----------------------PGAALAGSGDEGGADEFDAAFFLVQAQR
MAV_3570|M.avium_104 D--------------MADEAAKLLERFRAMEPWLQAGRVHAPFWEVHATR
MUL_3309|M.ulcerans_Agy99 E--------------VADQVGPLVERFKTIQPWLTDGIAHVPFWEVHATR
MAB_0868c|M.abscessus_ATCC_199 EQMPADMRERYQRPGVAEQAQQTFERLQQLD----DDMIHMPFFNIVAER
MLBr_00130|M.leprae_Br4923 -------------------REFIGVQGTQLSRYLEGGELSYRMYSFAKD-
:
Mflv_1324|M.gilvum_PYR-GCK EP-
TH_1722|M.thermoresistible__bu A--
Mvan_1888|M.vanbaalenii_PYR-1 L--
Mb1412c|M.bovis_AF2122/97 RGS
Rv1377c|M.tuberculosis_H37Rv RGS
MMAR_2193|M.marinum_M QN-
MSMEG_3041|M.smegmatis_MC2_155 R--
MAV_3570|M.avium_104 VD-
MUL_3309|M.ulcerans_Agy99 PD-
MAB_0868c|M.abscessus_ATCC_199 G--
MLBr_00130|M.leprae_Br4923 ---