For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MDLKIKDRVALVLASTAGLGAATARALAAEGAHVVITGRNAERAERLAASLPSATAVPVDLTEPRAVEHL LTATREAYGEPDIVVLNGPGPKPGAAADLDAEDIDTIGDLLLKTHVRLVKAVLPHMRAQGWGRILAIGSS GVQAPIPMLAASNIGRAGLAAYLKTLASEVAADGVTVNLQLPGRIATDRVAALDEAAAQRQGRSPDEVRA ASEASIPAGRYGTPDEFGALAAFLCSSAASYITGTAVRCDGGMLAHL
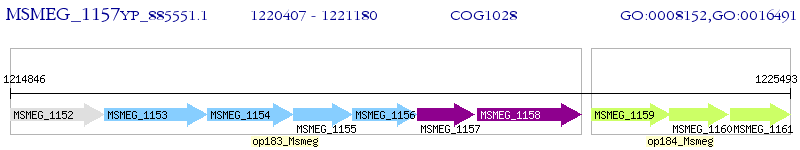
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1157 | - | - | 100% (257) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2918 | - | 7e-28 | 34.85% (264) | short-chain dehydrogenase/reductase SDR |
| M. smegmatis MC2 155 | MSMEG_3604 | - | 9e-28 | 36.19% (257) | sorbitol utilization protein SOU2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0950c | - | 1e-23 | 32.16% (255) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0296 | - | 2e-29 | 35.00% (260) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv0927c | - | 1e-23 | 32.16% (255) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 1e-19 | 27.53% (247) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_0833 | - | 2e-25 | 32.14% (252) | Short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_3383 | - | 1e-34 | 35.52% (259) | FabG-like dehydrogenase |
| M. avium 104 | MAV_2549 | - | 7e-31 | 35.36% (263) | short-chain dehydrogenase/reductase SDR |
| M. thermoresistible (build 8) | TH_3898 | - | 2e-27 | 35.29% (255) | PUTATIVE short chain dehydrogenase |
| M. ulcerans Agy99 | MUL_4442 | - | 3e-23 | 32.16% (255) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0045 | - | 2e-25 | 30.95% (252) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1157|M.smegmatis_MC2_155 --------------MDLKIKDRVALVLASTAGLGAATARALAAEGAHVVI
MMAR_3383|M.marinum_M --------------MDLRIAGRTALVTASSQGLGRACASALIAEGVSVVI
MAV_2549|M.avium_104 --------------MHYGIDGRSALVVGGSKGIGFEVAKLLAAEGARVAI
Mflv_0296|M.gilvum_PYR-GCK --------------MDMGLSGKRALVTGSSSGLGRAIAESLAAEGASVVV
MLBr_01807|M.leprae_Br4923 MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLAVARRLVADGHRVAV
Mvan_0045|M.vanbaalenii_PYR-1 --------------MSTDLRGRVALVTGAARGMGAAHARRLAAAGATVAV
Mb0950c|M.bovis_AF2122/97 -----------MILDMFRLDDKVAVITGGGRGLGAAIALAFAQAGADVLI
Rv0927c|M.tuberculosis_H37Rv -----------MILDMFRLDDKVAVITGGGRGLGAAIALAFAQAGADVLI
MUL_4442|M.ulcerans_Agy99 -----------MILDRFRLDDQVAVITGGGRGFGAAIALAFAEAGADVVI
TH_3898|M.thermoresistible__bu -----------VILDRFRLDDRVAVVTGAGRGLGAAMAVAFAEAGADVLI
MAB_0833|M.abscessus_ATCC_1997 -------MNRETFSRLFDLTGRTAIVTGGTRGIGRALAEGYACAGANVVV
: .: :: .. *:* * * * :
MSMEG_1157|M.smegmatis_MC2_155 TGRNAERAERLAASLPSATAVPVD----LTEPRAVEHLLTATREAYGEPD
MMAR_3383|M.marinum_M NGRETDKLERAAAEIGALATHGASC-RFVTADLTTDTGVQAIFDQVDQLD
MAV_2549|M.avium_104 LARTKTDVDAAVEAIRTEGGTAIGVTADVADDGQLTDAVREIAARHGAPL
Mflv_0296|M.gilvum_PYR-GCK HGRDEVRTGAVADGIRARGGDAWAA----VGDLATEGGAGAVAAAAGEVD
MLBr_01807|M.leprae_Br4923 THRA----SVVPDWFFGVEC-------DVTDNDAIGAAFKAVEEHQGPVE
Mvan_0045|M.vanbaalenii_PYR-1 NDRE----DSDALTALATEIGGLTAPGDVSDPKSCRALATHVAATTGRLD
Mb0950c|M.bovis_AF2122/97 ASRTSSELDAVAEQIRAAGRRAHTVAADLAHPEVTAQLAGQAVGAFGKLD
Rv0927c|M.tuberculosis_H37Rv ASRTSSELDAVAEQIRAAGRRAHTVAADLAHPEVTAQLAGQAVGAFGKLD
MUL_4442|M.ulcerans_Agy99 ASRTQSQLDEVADTVRSIGRRAHVVAADLAHPEATAELAGQAVEAFGRLD
TH_3898|M.thermoresistible__bu SARTQAQLEEVAAQIAATGRRAHVVAADLAHPEATAELAGAAVEAFGKLD
MAB_0833|M.abscessus_ATCC_1997 ASRKPEACTEAVERLRSLGARALGVPTHTGDIHSLEELVRATAEEFGGID
* .
MSMEG_1157|M.smegmatis_MC2_155 IVVLNGPGPKPGAAADLDAEDID-TIGDLLLKTHVRLVKAVLPHMRAQG-
MMAR_3383|M.marinum_M ILVTNNRGPRPADIIGVTDDDLA-ESLRLHYYVPIHLVQRYLPGMRDRR-
MAV_2549|M.avium_104 IVVGQAKYQRPGDFADITDVNVYRESFELHTMSQILLLHAVLPAMQDAG-
Mflv_0296|M.gilvum_PYR-GCK ILVNNAGVYDGLAWSDLAPRQWQ-SIYEVNVVSGVRMIEYLVPGMRQRG-
MLBr_01807|M.leprae_Br4923 VLVANAGITSDMLIMRMSEEQFQ-KVIDVNLTGVFRVAKLASRNMIKQR-
Mvan_0045|M.vanbaalenii_PYR-1 VLVANHAYMTMAPLLEHADDDWW-KVVDTNLGGTFFLVQSVLPHMRRTG-
Mb0950c|M.bovis_AF2122/97 IVVNNVGGTMPNTLLSTSTKDLA-DAFAFNVGTAHALTVAAVPLMLEHSG
Rv0927c|M.tuberculosis_H37Rv IVVNNVGGTMPNTLLSTSTKDLA-DAFAFNVGTAHALTVAAVPLMLEHSG
MUL_4442|M.ulcerans_Agy99 IVVNNVGGTMPNTLLTTSTKDLK-DAFTFNVATAHALTVAAVPLMLEHSG
TH_3898|M.thermoresistible__bu IVVNNVGGTMPGPLLNTSAKEMR-DAFTFNVGTAHALTTAAVPLMLKHSG
MAB_0833|M.abscessus_ATCC_1997 IVVNNAANALTEPVGAFTVDGWD-KSFGVNLRGPVFLVQAALPHLVASP-
::* : : :
MSMEG_1157|M.smegmatis_MC2_155 WGRILAIGSSGVQAPIP--MLAASNIGRAGLAAYLKTLASEVAADGVTVN
MMAR_3383|M.marinum_M FGRIVNITSEMVESPVA--GMIASAGARSALTTAMKGVQHDSVADNVTIN
MAV_2549|M.avium_104 WGRFVHIGSATAKEPAGNIHHAVANTSRPSTIGLLKTVSDEYAQFGITVN
Mflv_0296|M.gilvum_PYR-GCK WGRIIQIGGGLALQPAA--DQPHYNATLAARHNLTVSLARELAGTGVTSN
MLBr_01807|M.leprae_Br4923 FGRYIFMGSVVGLSGVP--GQANYAASKSGLIGMARSLARELGSRSITAN
Mvan_0045|M.vanbaalenii_PYR-1 AGRIVVIASEWGVTGWP--EATAYAAAKSGLISLVKTLGRELAREHIIVN
Mb0950c|M.bovis_AF2122/97 GGSVINISSTMGRLAAR--GFAAYGTAKAALAHYTRLAALDLCPR-VRVN
Rv0927c|M.tuberculosis_H37Rv GGSVINISSTMGRLAAR--GFAAYGTAKAALAHYTRLAALDLCPR-VRVN
MUL_4442|M.ulcerans_Agy99 GGGIINITSTMGRVAGR--GFAAYGTAKVALAHYTRLAALDLCPR-IRVN
TH_3898|M.thermoresistible__bu GGSIINITSTMGRLAGR--GFAAYGTAKAALAHYTRLAALDLAPR-IRVN
MAB_0833|M.abscessus_ATCC_1997 HAAVLNVVSVGAFMFGQ--GVSMYSAAKAALVSYTRSAAAELASRGVRVN
. : : . . : : *
MSMEG_1157|M.smegmatis_MC2_155 LQLPGRIATDRVAALDEAAAQRQGRSPDEVRAASEASIP-----AGRYGT
MMAR_3383|M.marinum_M QLLPMLIDSPRQLQMAQLLSESSGVAFEEIRRRQASVTS-----AKRLGR
MAV_2549|M.avium_104 TVAPGWIETENALAYLEQHLGASTDQERREFMLREARVP-----AARMGK
Mflv_0296|M.gilvum_PYR-GCK TVAPGAILTEPVREMTYRVAAQRGWGPTWEDVEKAAATTWFPNDIGRFGR
MLBr_01807|M.leprae_Br4923 VVAPGFIDT-----------EMTRAMDSKYQEAALQAIP-----LGRIGA
Mvan_0045|M.vanbaalenii_PYR-1 AVAPGVIDTPQ----LQVDADAAGVGLDDIHRRYAEAIP-----LGRIGS
Mb0950c|M.bovis_AF2122/97 AIAPGSILTS---------ALEVVAANDELRAPMEQATP-----LRRLGD
Rv0927c|M.tuberculosis_H37Rv AIAPGSILTS---------ALEVVAANDELRAPMEQATP-----LRRLGD
MUL_4442|M.ulcerans_Agy99 AIAPGSILTS---------ALEVVAANDELRAPMEQATP-----LRRLGD
TH_3898|M.thermoresistible__bu AIAPGSILTS---------ALDVVASNDELRTPMEQATP-----LRRLGE
MAB_0833|M.abscessus_ATCC_1997 ALAPGAIDTD---------MVRKTPPAEVER--MANTNH-----LKRLGL
* * : * *
MSMEG_1157|M.smegmatis_MC2_155 PDEFGALAAFLCSSAASYITGTAVRCDGG---MLAHL--------
MMAR_3383|M.marinum_M PEEVGAACAFLCSEQAGYISGVSLPVNGGGPVMGGGA--------
MAV_2549|M.avium_104 PSEIASLIAYLCSEPAGYLTGSWIEVDGG---LHRSAF-------
Mflv_0296|M.gilvum_PYR-GCK PEEVAAAVAFLAGVHAGYISGADLRVDGG--AIRSVA--------
MLBr_01807|M.leprae_Br4923 VEEIAGAVSFLASEDAGYISGAVIPVDGG---LGMGH--------
Mvan_0045|M.vanbaalenii_PYR-1 ADEVAVAVELLSDFDLEAVVGQVISCNGG---STRTRA-------
Mb0950c|M.bovis_AF2122/97 PVDIAAAAVYLASPAGSFLTGKTLEVDGG---LTFPNLDLPIPDL
Rv0927c|M.tuberculosis_H37Rv PVDIAAAAVYLASPAGSFLTGKTLEVDGG---LTFPNLDLPIPDL
MUL_4442|M.ulcerans_Agy99 PVDIAAAAVYLASPAGSFLTGKTLEVDGG---LTHPNLDIPVPDL
TH_3898|M.thermoresistible__bu PADIAAAAVYLASPAGSYLTGKVLEVDGG---LTRPNLDMPIPDL
MAB_0833|M.abscessus_ATCC_1997 PDEMVGTALLLTSDAGSYITGQTIIADGG---LVAR---------
:. * . : * : :**