For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MNRETFSRLFDLTGRTAIVTGGTRGIGRALAEGYACAGANVVVASRKPEACTEAVERLRSLGARALGVPT HTGDIHSLEELVRATAEEFGGIDIVVNNAANALTEPVGAFTVDGWDKSFGVNLRGPVFLVQAALPHLVAS PHAAVLNVVSVGAFMFGQGVSMYSAAKAALVSYTRSAAAELASRGVRVNALAPGAIDTDMVRKTPPAEVE RMANTNHLKRLGLPDEMVGTALLLTSDAGSYITGQTIIADGGLVAR
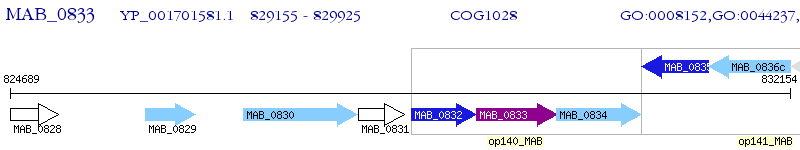
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0833 | - | - | 100% (256) | Short-chain dehydrogenase/reductase |
| M. abscessus ATCC 19977 | MAB_0164 | - | 3e-80 | 58.33% (252) | short chain dehydrogenase/reductase |
| M. abscessus ATCC 19977 | MAB_4632 | - | 4e-39 | 37.20% (250) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2788c | fabG | 3e-40 | 39.59% (245) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. gilvum PYR-GCK | Mflv_4818 | - | 1e-86 | 63.49% (252) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv2766c | fabG | 3e-40 | 39.59% (245) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. leprae Br4923 | MLBr_02565 | fabG | 6e-22 | 31.09% (238) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. marinum M | MMAR_1952 | fabG | 4e-37 | 37.96% (245) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_3655 | fabG | 2e-41 | 39.59% (245) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. smegmatis MC2 155 | MSMEG_1740 | - | 3e-85 | 61.51% (252) | dehydrogenase/reductase SDR family protein member 4 |
| M. thermoresistible (build 8) | TH_2589 | - | 2e-97 | 69.05% (252) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_2173 | fabG | 9e-37 | 37.55% (245) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. vanbaalenii PYR-1 | Mvan_1624 | - | 1e-87 | 63.89% (252) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4818|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1624|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1740|M.smegmatis_MC2_155 --------------------------------------------------
TH_2589|M.thermoresistible__bu --------------------------------------------------
MAB_0833|M.abscessus_ATCC_1997 --------------------------------------------------
Mb2788c|M.bovis_AF2122/97 --------------------------------------------------
Rv2766c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1952|M.marinum_M --------------------------------------------------
MUL_2173|M.ulcerans_Agy99 --------------------------------------------------
MAV_3655|M.avium_104 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 MAPKCSSDLFSQIVSSGPGSFVAKQLGVPRPETLRRYRPSDAPLCGSLLI
Mflv_4818|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1624|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1740|M.smegmatis_MC2_155 --------------------------------------------------
TH_2589|M.thermoresistible__bu --------------------------------------------------
MAB_0833|M.abscessus_ATCC_1997 --------------------------------------------------
Mb2788c|M.bovis_AF2122/97 --------------------------------------------------
Rv2766c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1952|M.marinum_M --------------------------------------------------
MUL_2173|M.ulcerans_Agy99 --------------------------------------------------
MAV_3655|M.avium_104 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 GGDGRLVEPLRATLEKDYDLVSNNLGGRWTDSFGGLVFDATGITAPAELK
Mflv_4818|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1624|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1740|M.smegmatis_MC2_155 --------------------------------------------------
TH_2589|M.thermoresistible__bu --------------------------------------------------
MAB_0833|M.abscessus_ATCC_1997 --------------------------------------------------
Mb2788c|M.bovis_AF2122/97 --------------------------------------------------
Rv2766c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1952|M.marinum_M --------------------------------------------------
MUL_2173|M.ulcerans_Agy99 --------------------------------------------------
MAV_3655|M.avium_104 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 GLHNFFTPLLRNLGRCARVVVIGTTPDATTSTDERIAQRALEGFSRSLGK
Mflv_4818|M.gilvum_PYR-GCK -----------------------------------------------MDR
Mvan_1624|M.vanbaalenii_PYR-1 -----------------------------------------------MDR
MSMEG_1740|M.smegmatis_MC2_155 -----------------------------------------------MDR
TH_2589|M.thermoresistible__bu -----------------------------------------------VDR
MAB_0833|M.abscessus_ATCC_1997 -----------------------------------------------MNR
Mb2788c|M.bovis_AF2122/97 --------------------------------------------------
Rv2766c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1952|M.marinum_M --------------------------------------------------
MUL_2173|M.ulcerans_Agy99 --------------------------------------------------
MAV_3655|M.avium_104 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 ELRRGATVALVYLSPDAKPAATGLESTMRFILSAKSAYVNGQVFYVGAAD
Mflv_4818|M.gilvum_PYR-GCK ASFDRLFD--MTDRTVVVTGGTRGIGLALAEGYALAGARVVVASRK--AD
Mvan_1624|M.vanbaalenii_PYR-1 ASFDRLFD--MTDRTVVVTGGTRGIGLALAEGYVLAGARVVVASRK--AD
MSMEG_1740|M.smegmatis_MC2_155 QRFERLFD--MSDRTVIVTGGTRGIGLSLAEGFVLAGARVVVASRK--PD
TH_2589|M.thermoresistible__bu QTFDKLFD--MTGRTVVVTGGTRGIGLALAEGYVLAGARVVVASRK--SD
MAB_0833|M.abscessus_ATCC_1997 ETFSRLFD--LTGRTAIVTGGTRGIGRALAEGYACAGANVVVASRK--PE
Mb2788c|M.bovis_AF2122/97 ---MTSLD--LTGRTAIITGASRGIGLAIAQQLAAAGAHVVLTARR--QE
Rv2766c|M.tuberculosis_H37Rv ---MTSLD--LTGRTAIITGASRGIGLAIAQQLAAAGAHVVLTARR--QE
MMAR_1952|M.marinum_M ---MTSLD--LTGRTAIITGASRGIGLAIAQRLAGAGANVVLTARK--QE
MUL_2173|M.ulcerans_Agy99 ---MTSLD--LTGRTAIITGASRGIGLAIAQRLAGAGANVVLTARK--QE
MAV_3655|M.avium_104 ---MTSQD--LTGRTAIITGASRGIGLAIAQQLAAAGANVVLTARK--QE
MLBr_02565|M.leprae_Br4923 STPPTNWDRPLENKVAIVTGAARGIGAAIAEVVARDGARVVAIDVESAAE
* : .:..::**.:**** ::*: . **.** . :
Mflv_4818|M.gilvum_PYR-GCK ACEQAAHHLRELGAQAIGVPTHLGQVDDLGALVERTVAEFGGLDVLVNNA
Mvan_1624|M.vanbaalenii_PYR-1 ACEETARHLRELGGSAIGVPTHLGEVDDLGALVERTVAEFGGVDVLVNNA
MSMEG_1740|M.smegmatis_MC2_155 ACETAAAHLRELGGDAVGVPAHTGDVDDLGAVVERAVSEFGGVDVVVNNA
TH_2589|M.thermoresistible__bu ACERAAAHLRALGGDAIGVPTHLGDIDACEALVRRTVDEFGGIDVLVNNA
MAB_0833|M.abscessus_ATCC_1997 ACTEAVERLRSLGARALGVPTHTGDIHSLEELVRATAEEFGGIDIVVNNA
Mb2788c|M.bovis_AF2122/97 AADEAAAQV---GDRALGVGAHAVDEDAARRCVDLTLERFGSVDILINNA
Rv2766c|M.tuberculosis_H37Rv AADEAAAQV---GDRALGVGAHAVDEDAARRCVDLTLERFGSVDILINNA
MMAR_1952|M.marinum_M SADAAAAEV---GERAVGVGAHAVDEEAARGCVDLALERFGSVDILINNA
MUL_2173|M.ulcerans_Agy99 SADAAAAEV---GERAVGVGAHAVDEEAARGCVDLALERFGSVDILINNA
MAV_3655|M.avium_104 AADEAAAQV---GPQALGVGAHAVDEDAARQCVQLTLERFGSVDILVNNA
MLBr_02565|M.leprae_Br4923 ALDETANRVG-----GTTLSLDVTADDAVDKITEHLHDYQGGHADILVNN
: :. .: . : . . . *. :: *
Mflv_4818|M.gilvum_PYR-GCK AN-PLAQPFGHMTVDALSKSFEVNLQGPVMLTQEALPHLKASDHAAVLNM
Mvan_1624|M.vanbaalenii_PYR-1 AN-PLAQPFGYMTVDALTKSFEVNLRGPVMLVQEALPHLKASEHAAVLNM
MSMEG_1740|M.smegmatis_MC2_155 AN-ALAQPLGKMTPEAWAKSYEVNLRGPVFLVQHALPYLQKSQNAAVLNM
TH_2589|M.thermoresistible__bu AN-ALTQPLGELTVDAWTKSFEVNLRGPVFLVQAALPHLKTSPHAAVLNM
MAB_0833|M.abscessus_ATCC_1997 AN-ALTEPVGAFTVDGWDKSFGVNLRGPVFLVQAALPHLVASPHAAVLNV
Mb2788c|M.bovis_AF2122/97 GTNPAYGPLLEQDHARFAKIFDVNLWAPLMWTSLVVTAWMGEHGGAVVNT
Rv2766c|M.tuberculosis_H37Rv GTNPAYGPLLEQDHARFAKIFDVNLWAPLMWTSLVVTAWMGEHGGAVVNT
MMAR_1952|M.marinum_M GTNPAYGPLIEQDHARFTKIFDVNLWAPLMWTSLVVKAWMGEHGGVVINT
MUL_2173|M.ulcerans_Agy99 GTNPAYGPLIEQDHARFTKIFDVNLWAPLMWTSLVVKAWMGEHGGVVINT
MAV_3655|M.avium_104 GTNPAYGPLIEQDHARFAKIFDVNLWAPLLWTSLAVKSWMGEHGGSIVNT
MLBr_02565|M.leprae_Br4923 AGITRDKLLANMDDCRWDAVLEVNLLAPQRLTEGLVGNGSISKGGRVIVL
. . . *** .* .. : . . ::
Mflv_4818|M.gilvum_PYR-GCK VSVGAFIFAPMLSIYASMKAAMMSFTRSMAAEFVHDGIRVNALAPGPVDT
Mvan_1624|M.vanbaalenii_PYR-1 VSVGAFIFAPMLSIYASMKAAMMSFTRSMAAEFVHHGIRVNALAPGPVDT
MSMEG_1740|M.smegmatis_MC2_155 VSVGAFNFAPALSIYASSKAALMSVTRSMAAEFAPSGIRVNALAPGPVDT
TH_2589|M.thermoresistible__bu VSVGAFLFSPGTAMYSAGKAALMSFTRSMAAQFAPLGIRVNALAPGPVDT
MAB_0833|M.abscessus_ATCC_1997 VSVGAFMFGQGVSMYSAAKAALVSYTRSAAAELASRGVRVNALAPGAIDT
Mb2788c|M.bovis_AF2122/97 ASIGGMHQSPAMGMYNATKAALIHVTKQLALELSPR-IRVNAICPGVVRT
Rv2766c|M.tuberculosis_H37Rv ASIGGMHQSPAMGMYNATKAALIHVTKQLALELSPR-IRVNAICPGVVRT
MMAR_1952|M.marinum_M ASIGGMHQSPAMGMYNATKAALIHVTKQLALELSPR-VRVNAICPGVVRT
MUL_2173|M.ulcerans_Agy99 ASIGGMHQSPAMGMYNATNAALIHVTKQLALELSPR-VRVNAICPGVVRT
MAV_3655|M.avium_104 ASIGGLHQSPAMGMYNATKAALIHVTKQLALELSPR-VRVNAIAPGVVRT
MLBr_02565|M.leprae_Br4923 SSMAGIAGNRGQTNYATAKAGVIGLTQALAPGLAEKGITINAVAPGFIET
*:..: * : :*.:: *: * : : :**:.** : *
Mflv_4818|M.gilvum_PYR-GCK DMMRKNPQEAIDGMVGGTLVGRLASADEMVGAALLLTSDAGSYITGQVII
Mvan_1624|M.vanbaalenii_PYR-1 DMMRKNPQEAIDGMVGGTLLGRLATPDEMVGAALLLTSDAGSYITGQVII
MSMEG_1740|M.smegmatis_MC2_155 DMMRKNPQEAIDAMTAGTLMKRLAHPDEMVGAALLLCSDAGSYMTGQVVI
TH_2589|M.thermoresistible__bu DMMRNNPQEVVDMMAGATLMKRLATPDEMVGAALLLTSDAGSYITGTVLI
MAB_0833|M.abscessus_ATCC_1997 DMVRKTPPAEVERMANTNHLKRLGLPDEMVGTALLLTSDAGSYITGQTII
Mb2788c|M.bovis_AF2122/97 RLAEALWKDHEDPLAATIALGRIGEPADIASAVAFLVSDAASWITGETMI
Rv2766c|M.tuberculosis_H37Rv RLAEALWKDHEDPLAATIALGRIGEPADIASAVAFLVSDAASWITGETMI
MMAR_1952|M.marinum_M KLAEALWKDHEDPLAANIALGRIGEPVDVAEAVGFLVSDAASWITGETMV
MUL_2173|M.ulcerans_Agy99 KLAEALWKDHEDPLAANIALGRIGEPVDVAEAVGFLVSDAASWITGETMV
MAV_3655|M.avium_104 RLAEALWKDHEDPLSSSIALGRIGEPIDVAAAVAFLVSDAASWITGETLV
MLBr_02565|M.leprae_Br4923 QMTAAIPLATREVGRRLNSLLQGGLPVDVAETIAYFAIPASNAVTGNVIR
: : : : . . ::. : : *.. :** .:
Mflv_4818|M.gilvum_PYR-GCK ADGGGTPR-------------
Mvan_1624|M.vanbaalenii_PYR-1 ADGGGTPR-------------
MSMEG_1740|M.smegmatis_MC2_155 VDGGGTPR-------------
TH_2589|M.thermoresistible__bu ADGGGTPR-------------
MAB_0833|M.abscessus_ATCC_1997 ADGGLVAR-------------
Mb2788c|M.bovis_AF2122/97 IDGGLLLGNALGFRAAPSTEH
Rv2766c|M.tuberculosis_H37Rv IDGGLLLGNALGFRAAPSTEH
MMAR_1952|M.marinum_M IDGGLLLGSAAGFQSRPGSAS
MUL_2173|M.ulcerans_Agy99 IDGGLLLGSAAGFQSRPGSAS
MAV_3655|M.avium_104 IDGGTVLGAAQGFRSAPTPS-
MLBr_02565|M.leprae_Br4923 VCGQAMLGA------------
*