For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ILDMFRLDDKVAVITGGGRGLGAAIALAFAQAGADVLIASRTSSELDAVAEQIRAAGRRAHTVAADLAHP EVTAQLAGQAVGAFGKLDIVVNNVGGTMPNTLLSTSTKDLADAFAFNVGTAHALTVAAVPLMLEHSGGGS VINISSTMGRLAARGFAAYGTAKAALAHYTRLAALDLCPRVRVNAIAPGSILTSALEVVAANDELRAPME QATPLRRLGDPVDIAAAAVYLASPAGSFLTGKTLEVDGGLTFPNLDLPIPDL*
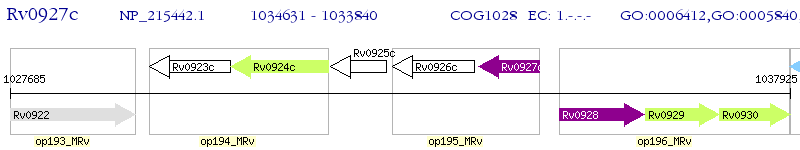
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0927c | - | - | 100% (263) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv1928c | - | 2e-35 | 33.98% (256) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv2766c | fabG | 2e-34 | 38.34% (253) | 3-ketoacyl-(acyl-carrier-protein) reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0950c | - | 1e-143 | 100.00% (263) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1825 | - | 1e-121 | 82.13% (263) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_02565 | fabG | 2e-19 | 33.06% (245) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. abscessus ATCC 19977 | MAB_4632 | - | 4e-97 | 70.34% (263) | short chain dehydrogenase |
| M. marinum M | MMAR_4581 | - | 1e-130 | 88.59% (263) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_1046 | - | 1e-130 | 87.83% (263) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5584 | - | 1e-120 | 81.75% (263) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_3898 | - | 1e-121 | 82.13% (263) | PUTATIVE short chain dehydrogenase |
| M. ulcerans Agy99 | MUL_4442 | - | 1e-129 | 87.45% (263) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_4921 | - | 1e-122 | 83.65% (263) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Rv0927c|M.tuberculosis_H37Rv --------------------------------------------------
Mb0950c|M.bovis_AF2122/97 --------------------------------------------------
MMAR_4581|M.marinum_M --------------------------------------------------
MUL_4442|M.ulcerans_Agy99 --------------------------------------------------
MAV_1046|M.avium_104 --------------------------------------------------
Mflv_1825|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4921|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3898|M.thermoresistible__bu --------------------------------------------------
MSMEG_5584|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4632|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 MAPKCSSDLFSQIVSSGPGSFVAKQLGVPRPETLRRYRPSDAPLCGSLLI
Rv0927c|M.tuberculosis_H37Rv --------------------------------------------------
Mb0950c|M.bovis_AF2122/97 --------------------------------------------------
MMAR_4581|M.marinum_M --------------------------------------------------
MUL_4442|M.ulcerans_Agy99 --------------------------------------------------
MAV_1046|M.avium_104 --------------------------------------------------
Mflv_1825|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4921|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3898|M.thermoresistible__bu --------------------------------------------------
MSMEG_5584|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4632|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 GGDGRLVEPLRATLEKDYDLVSNNLGGRWTDSFGGLVFDATGITAPAELK
Rv0927c|M.tuberculosis_H37Rv --------------------------------------MILDMFR-----
Mb0950c|M.bovis_AF2122/97 --------------------------------------MILDMFR-----
MMAR_4581|M.marinum_M --------------------------------------MILDRFR-----
MUL_4442|M.ulcerans_Agy99 --------------------------------------MILDRFR-----
MAV_1046|M.avium_104 --------------------------------------MILDRFR-----
Mflv_1825|M.gilvum_PYR-GCK --------------------------------------MILDRFR-----
Mvan_4921|M.vanbaalenii_PYR-1 ----------------------------MTRGRQREANVILDRFR-----
TH_3898|M.thermoresistible__bu --------------------------------------VILDRFR-----
MSMEG_5584|M.smegmatis_MC2_155 -------------------------------------MLILDRFR-----
MAB_4632|M.abscessus_ATCC_1997 --------------------------------------MILDRFK-----
MLBr_02565|M.leprae_Br4923 GLHNFFTPLLRNLGRCARVVVIGTTPDATTSTDERIAQRALEGFSRSLGK
*: *
Rv0927c|M.tuberculosis_H37Rv --------------------------------------------------
Mb0950c|M.bovis_AF2122/97 --------------------------------------------------
MMAR_4581|M.marinum_M --------------------------------------------------
MUL_4442|M.ulcerans_Agy99 --------------------------------------------------
MAV_1046|M.avium_104 --------------------------------------------------
Mflv_1825|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4921|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3898|M.thermoresistible__bu --------------------------------------------------
MSMEG_5584|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4632|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 ELRRGATVALVYLSPDAKPAATGLESTMRFILSAKSAYVNGQVFYVGAAD
Rv0927c|M.tuberculosis_H37Rv ----------LDDKVAVITGGGRGLGAAIALAFAQAGADVLIASRTSSEL
Mb0950c|M.bovis_AF2122/97 ----------LDDKVAVITGGGRGLGAAIALAFAQAGADVLIASRTSSEL
MMAR_4581|M.marinum_M ----------LDDQVAVITGGGRGLGAAIALAFAEAGADVVIASRTQSQL
MUL_4442|M.ulcerans_Agy99 ----------LDDQVAVITGGGRGFGAAIALAFAEAGADVVIASRTQSQL
MAV_1046|M.avium_104 ----------LDDKVAVITGAGRGLGAAIAVAFAEAGADVLIASRTESQL
Mflv_1825|M.gilvum_PYR-GCK ----------LDDQVAVVTGAGRGLGAAMALAFAEAGADVLIAARTQSQL
Mvan_4921|M.vanbaalenii_PYR-1 ----------LDDQVAVVTGAGRGLGAAMALAFAEAGADVLIAARTRSQL
TH_3898|M.thermoresistible__bu ----------LDDRVAVVTGAGRGLGAAMAVAFAEAGADVLISARTQAQL
MSMEG_5584|M.smegmatis_MC2_155 ----------LDDKVAVVTGAGRGLGAAIALAFAEAGADVVIAARTQSQL
MAB_4632|M.abscessus_ATCC_1997 ----------LDGNVAVVTGAGRGLGAAIAEAFAQAGADVLIASRTESQL
MLBr_02565|M.leprae_Br4923 STPPTNWDRPLENKVAIVTGAARGIGAAIAEVVARDGARVVAID-VESAA
*:..**::**..**:***:* ..*. ** *: . :
Rv0927c|M.tuberculosis_H37Rv DAVAEQIRAAGRRAHTVAADLAHPEVTAQLAGQAVGAFG-KLDIVVNNVG
Mb0950c|M.bovis_AF2122/97 DAVAEQIRAAGRRAHTVAADLAHPEVTAQLAGQAVGAFG-KLDIVVNNVG
MMAR_4581|M.marinum_M DEVADTVRSIGRRAHVVAADLAHPEATAELAGQAVEAFG-RLDIVVNNVG
MUL_4442|M.ulcerans_Agy99 DEVADTVRSIGRRAHVVAADLAHPEATAELAGQAVEAFG-RLDIVVNNVG
MAV_1046|M.avium_104 EAVAEQVRAAGRRAHVVAADLAHPESTAELAGRAVEAFG-KLDIVVNNVG
Mflv_1825|M.gilvum_PYR-GCK EAVAEQIAGTGRRAHIVVADLARPEDTAGLAAQAVEAFG-RLDIVVNNVG
Mvan_4921|M.vanbaalenii_PYR-1 EEVAEQIEGTGRRAHIVVADLAHAEDTAALAAGAVEAFG-RLDIVVNNVG
TH_3898|M.thermoresistible__bu EEVAAQIAATGRRAHVVAADLAHPEATAELAGAAVEAFG-KLDIVVNNVG
MSMEG_5584|M.smegmatis_MC2_155 DEVAGQVAGAGRKAHVVVADLAHPDATAELVQAAIEAFG-KLDVVVNNVG
MAB_4632|M.abscessus_ATCC_1997 REVAAKVQAAGRQAEVVVADLSDTEASAALAQKAVDRFG-RLDVVVNNVG
MLBr_02565|M.leprae_Br4923 EALDETANRVG--GTTLSLDVTADDAVDKITEHLHDYQGGHADILVNNAG
: * . : *:: : :. * : *::***.*
Rv0927c|M.tuberculosis_H37Rv GTMPNTLLSTSTKDLADAFAFNVGTAHALTVAAVPLMLEHSGGGSVINIS
Mb0950c|M.bovis_AF2122/97 GTMPNTLLSTSTKDLADAFAFNVGTAHALTVAAVPLMLEHSGGGSVINIS
MMAR_4581|M.marinum_M GTMPNTLLTTSTKDLKDAFTFNVATAHALTVAAVPLMLEHSGGGSIINIT
MUL_4442|M.ulcerans_Agy99 GTMPNTLLTTSTKDLKDAFTFNVATAHALTVAAVPLMLEHSGGGGIINIT
MAV_1046|M.avium_104 GTMPNTLLTTSTKDLKDAFTFNVATAHALTVAAVPLMLEHSGGGNIINIT
Mflv_1825|M.gilvum_PYR-GCK GTMPNALLNTSTKDMRDAFTFNVATAHALTTAAVPLMLEHSGGGSIINIT
Mvan_4921|M.vanbaalenii_PYR-1 GTMPNALLTTSTKDMRDAFAFNVATAHALTTAAVPLMLEHSGGGSIINIT
TH_3898|M.thermoresistible__bu GTMPGPLLNTSAKEMRDAFTFNVGTAHALTTAAVPLMLKHSGGGSIINIT
MSMEG_5584|M.smegmatis_MC2_155 GTMPAPLLNTSTKDLRDAFTFNVSTAHALTSAAVPLMLEHSGGGSIINIT
MAB_4632|M.abscessus_ATCC_1997 GTMPKPFLDTTNDDLAEAFSFNVLSGHALLRAAVPHLLK-SDNASVINIT
MLBr_02565|M.leprae_Br4923 ITRDKLLANMDDCRWDAVLEVNLLAPQRLTEGLVGNGSISKGGR-VIVLS
* : .: .*: : : * . * ... :* ::
Rv0927c|M.tuberculosis_H37Rv STMGRLAARGFAAYGTAKAALAHYTRLAALDLCPR-VRVNAIAPGSILTS
Mb0950c|M.bovis_AF2122/97 STMGRLAARGFAAYGTAKAALAHYTRLAALDLCPR-VRVNAIAPGSILTS
MMAR_4581|M.marinum_M STMGRVAGRGFAAYGTAKAALAHYTRLAALDLCPR-IRVNAIAPGSILTS
MUL_4442|M.ulcerans_Agy99 STMGRVAGRGFAAYGTAKVALAHYTRLAALDLCPR-IRVNAIAPGSILTS
MAV_1046|M.avium_104 STMGRLAGRGFAAYGTAKAALSHYTRLTALDLCPR-IRVNAIAPGSILTS
Mflv_1825|M.gilvum_PYR-GCK STMGRVAGRGFAAYGTAKAALAHYTRLSALDLAPR-IRVNAIAPGSILTS
Mvan_4921|M.vanbaalenii_PYR-1 STMGRLAGRGFAAYGTAKAALAHYTRLSALDLAPR-IRVNAIAPGSILTS
TH_3898|M.thermoresistible__bu STMGRLAGRGFAAYGTAKAALAHYTRLAALDLAPR-IRVNAIAPGSILTS
MSMEG_5584|M.smegmatis_MC2_155 STMGRLAGRGFVAYGTAKAALAHYTRLAALDLCPR-IRVNAIAPGSILTS
MAB_4632|M.abscessus_ATCC_1997 STMGRLPGRAFLGYCTAKGALAHYTRTAAMDLSPR-IRVNGIAPGSILTS
MLBr_02565|M.leprae_Br4923 SMAGIAGNRGQTNYATAKAGVIGLTQALAPGLAEKGITINAVAPGFIETQ
* * *. * *** .: *: * .*. : : :*.:*** * *.
Rv0927c|M.tuberculosis_H37Rv ALEVVAANDELRAPMEQATPLRRLGDPVDIAAAAVYLASPAGSFLTGKTL
Mb0950c|M.bovis_AF2122/97 ALEVVAANDELRAPMEQATPLRRLGDPVDIAAAAVYLASPAGSFLTGKTL
MMAR_4581|M.marinum_M ALEVVAANDELRAPMEQATPLRRLGDPVDIAAAAVYLASPAGSFLTGKTL
MUL_4442|M.ulcerans_Agy99 ALEVVAANDELRAPMEQATPLRRLGDPVDIAAAAVYLASPAGSFLTGKTL
MAV_1046|M.avium_104 ALDVVASNDELRAPMEKATPLRRLGDPVDIASAAVYLASPAGEFLTGKTL
Mflv_1825|M.gilvum_PYR-GCK ALEIVASNDDLRDPMEKATPMKRLGDPLDIAAAAVYLASPAGSYLTGKTL
Mvan_4921|M.vanbaalenii_PYR-1 ALEIVADNDELRGPMEKATPLRRLGDPLDIAAAAVYLASPAGSYLTGKTL
TH_3898|M.thermoresistible__bu ALDVVASNDELRTPMEQATPLRRLGEPADIAAAAVYLASPAGSYLTGKVL
MSMEG_5584|M.smegmatis_MC2_155 ALDVVASNDALREPMEKVTPLRRLGDPADIAAAAVYLASPAGAYLTGKTL
MAB_4632|M.abscessus_ATCC_1997 ALEVVAGNDEMRNTLEQNTPLHRLGDPADIAAAAVYLASPAGSFLTGKVL
MLBr_02565|M.leprae_Br4923 MTAAIP--LATREVGRRLNSLLQGGLPVDVAETIAYFAIPASNAVTGNVI
:. * .: ..: : * * *:* : .*:* **. :**:.:
Rv0927c|M.tuberculosis_H37Rv EVDGGLTFPNLDLPIPDL-
Mb0950c|M.bovis_AF2122/97 EVDGGLTFPNLDLPIPDL-
MMAR_4581|M.marinum_M EVDGGLTHPNLDIPVPDL-
MUL_4442|M.ulcerans_Agy99 EVDGGLTHPNLDIPVPDL-
MAV_1046|M.avium_104 EVDGGLTYPNLDIPVPDL-
Mflv_1825|M.gilvum_PYR-GCK EVDGGITFPNLELPIPDL-
Mvan_4921|M.vanbaalenii_PYR-1 EVDGGITFPNLDLPIPDL-
TH_3898|M.thermoresistible__bu EVDGGLTRPNLDMPIPDL-
MSMEG_5584|M.smegmatis_MC2_155 EVDGGLTMPNLDLPIPDL-
MAB_4632|M.abscessus_ATCC_1997 EVDGGLIAPNLDIPLPDLA
MLBr_02565|M.leprae_Br4923 RVCGQAMLGA---------
.* *