For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DVRSAFDLSGRTALVTGGNQGLGKAFAIALAQAGARVSFSGRNAERNEKTAAEAAAAGHQLHAITADITR AEDVERMTAEAIEALGHIDILVNNAGTCHHGESWTVTEEQWDDVFDLNVKALWACSLAVGAHMRERGSGS VVNIGSMSGIIVNRPQMQPAYNASKAAVHHLTKSLAAEWAPLGIRVNALAPGYVKTDMAPVDRPEFKRYW IDDTPQLRYAVPEEIAPSVVFLASDAASFITGSVLVADGGYTAW*
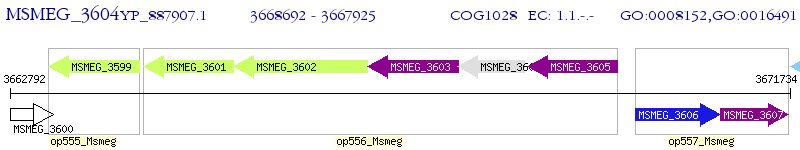
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3604 | - | - | 100% (255) | sorbitol utilization protein SOU2 |
| M. smegmatis MC2 155 | MSMEG_5204 | - | 7e-43 | 43.37% (249) | oxidoreductase, short chain dehydrogenase/reductase family |
| M. smegmatis MC2 155 | MSMEG_3420 | - | 3e-40 | 35.94% (256) | gluconate 5-dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1963c | - | 2e-43 | 38.98% (254) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1263 | - | 7e-42 | 38.89% (252) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv1928c | - | 2e-43 | 38.98% (254) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 1e-23 | 29.58% (240) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_4632 | - | 1e-33 | 35.20% (250) | short chain dehydrogenase |
| M. marinum M | MMAR_2849 | - | 7e-43 | 38.58% (254) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_2775 | - | 6e-43 | 40.71% (253) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_4266 | - | 8e-41 | 37.30% (252) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_3898 | fabG | 6e-38 | 37.97% (237) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. vanbaalenii PYR-1 | Mvan_0894 | - | 2e-43 | 40.87% (252) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1263|M.gilvum_PYR-GCK -------MSDR-----------------FSMEGRVAVITGGGTGIGRASA
TH_4266|M.thermoresistible__bu -------VDDR-----------------FSMAGRVAVITGGGTGIGRASA
MAB_4632|M.abscessus_ATCC_1997 ------MILDR-----------------FKLDGNVAVVTGAGRGLGAAIA
Mb1963c|M.bovis_AF2122/97 ---------MSVLDL-------------FDLHGKRALITGASTGIGKRVA
Rv1928c|M.tuberculosis_H37Rv ---------MSVLDL-------------FDLHGKRALITGASTGIGKRVA
MMAR_2849|M.marinum_M ---------MSVLDL-------------FDLSGKRALITGASTGIGKKVA
MAV_2775|M.avium_104 -----------MLDL-------------FDLRGRRALVTGASTGIGKQVA
MSMEG_3604|M.smegmatis_MC2_155 ---------MDVRSA-------------FDLSGRTALVTGGNQGLGKAFA
Mvan_0894|M.vanbaalenii_PYR-1 ---------MTVDIAREQPATAPRPDQMFDLTGRTALVTGASSGLGVQFA
MLBr_01807|M.leprae_Br4923 MTDAAAKVSAAPGRGK------------PAFVSRSILVTGGNRGIGLAVA
MUL_3898|M.ulcerans_Agy99 ------MCGAHRDRG-----------------YVYPVVTGGAQGLGLAIA
::**. *:* *
Mflv_1263|M.gilvum_PYR-GCK LVLAEHGADVVLAGRRAEPLKETAAEVEALG-RRALAVPTDVTDADACQA
TH_4266|M.thermoresistible__bu LVLAEYGADIVLAGRRPEPLKKTAAEIEALG-RRALAVPTDVTAPDECRA
MAB_4632|M.abscessus_ATCC_1997 EAFAQAGADVLIASRTESQLREVAAKVQAAG-RQAEVVVADLSDTEASAA
Mb1963c|M.bovis_AF2122/97 LAYVEAGAQVAIAARHLDALEKLADEIGTSG-GKVVPVCCDVSQHQQVTS
Rv1928c|M.tuberculosis_H37Rv LAYVEAGAQVAIAARHLDALEKLADEIGTSG-GKVVPVCCDVSQHQQVTS
MMAR_2849|M.marinum_M LAYAEAGAQVAVAARHSDALQVVADEIAGVG-GKALPIRCDVTQPDQVRG
MAV_2775|M.avium_104 QAYLQAGARVAIAARDFEALQRTADELTAGGTGGVVAIRCDVTQPDEVSG
MSMEG_3604|M.smegmatis_MC2_155 IALAQAGARVSFSGRNAERNEKTAAEAAAAG-HQLHAITADITRAEDVER
Mvan_0894|M.vanbaalenii_PYR-1 LALRSAGAEVVVAARRADR----LAELCEAH-EGLSAVSCDVADPASIES
MLBr_01807|M.leprae_Br4923 RRLVADGHRVAVTHR---ASVVPDWFFG---------VECDVTDNDAIGA
MUL_3898|M.ulcerans_Agy99 ERFVSEGARVVLGDVNLEATEAAAKQLGGGE--VAVAVRCDVTKADEVET
* : . : *::
Mflv_1263|M.gilvum_PYR-GCK LVDTTLAEFGRLDVLLNNAGGGETKSLMKWTDDEWHHVLDLNLSSAWYLS
TH_4266|M.thermoresistible__bu LVDATVGEFGRLDVLVNNAGGGATKPLMKWTDDEWHEVLNLNLSSAWYLS
MAB_4632|M.abscessus_ATCC_1997 LAQKAVDRFGRLDVVVNNVGGTMPKPFLDTTNDDLAEAFSFNVLSGHALL
Mb1963c|M.bovis_AF2122/97 MLDQVTAELGGIDIAVCNAGIITVTPMLDMPLEEFQRLQNTNVTGVFLTA
Rv1928c|M.tuberculosis_H37Rv MLDQVTAELGGIDIAVCNAGIITVTPMLDMPLEEFQRLQNTNVTGVFLTA
MMAR_2849|M.marinum_M MLDQMTGELGGIDIAVCNAGIVSVQAMLDMPLEEFQRIQDTNVTGVFLTA
MAV_2775|M.avium_104 MVDRMIAELGGIDIAVCNAGIISVSPMLEMPAAEFQRIQDTNVTGVFLTA
MSMEG_3604|M.smegmatis_MC2_155 MTAEAIEALGHIDILVNNAGTCHHGESWTVTEEQWDDVFDLNVKALWACS
Mvan_0894|M.vanbaalenii_PYR-1 AYQCVEERVGAVDILVNNAGTSIPARAEDESLTDFQYVLDVNLVGAFRLT
MLBr_01807|M.leprae_Br4923 AFKAVEEHQGPVEVLVANAGITSDMLIMRMSEEQFQKVIDVNLTGVFRVA
MUL_3898|M.ulcerans_Agy99 LLQAALERFGGLDIMVNNAGITRDATMRKMTEEQFDQVIDVHLKGTWNGI
* ::: : *.* . : . :: .
Mflv_1263|M.gilvum_PYR-GCK RAAAKPMIAQGSG-AIVNISSGAGLLAMP--QAPIYGAAKAGLQNLTGSM
TH_4266|M.thermoresistible__bu RAAAKPMIDQGRG-AIVNISSGAGLLAMP--QAPVYAAAKAGLNNLTGSM
MAB_4632|M.abscessus_ATCC_1997 RAAVPHLLKSDNA-SVINITSTMGRLPGR--AFLGYCTAKGALAHYTRTA
Mb1963c|M.bovis_AF2122/97 QAAAKAMVKQGQGGVIINTASMSGHIINVPQQVSHYCASKAAVIHLTKAM
Rv1928c|M.tuberculosis_H37Rv QAAAKAMVKQGQGGVIINTASMSGHIINVPQQVSHYCASKAAVIHLTKAM
MMAR_2849|M.marinum_M QAAARAMVDQGLGGTIITTASMSGHIINIPQQVSHYCTSKAAVVHLTKAM
MAV_2775|M.avium_104 QAAARAMVRQGRGGAIITTASMSGHIINVPQQVGHYCASKAAVIQLTKAM
MSMEG_3604|M.smegmatis_MC2_155 LAVGAHMRERGSG-SVVNIGSMSGIIVNRPQMQPAYNASKAAVHHLTKSL
Mvan_0894|M.vanbaalenii_PYR-1 QLVGRKMIERGEG-SIVNVASILGMVAAAPNRQAAYCASKGGLINLTREL
MLBr_01807|M.leprae_Br4923 KLASRNMIKQRFG-RYIFMGSVVGLSGVPG--QANYAASKSGLIGMARSL
MUL_3898|M.ulcerans_Agy99 RLAAAIMRENKRG-AIVNMSSVSGKVGMVG--QTNYSAAKAGIVGMTKAA
. : . : * * * ::*..: :
Mflv_1263|M.gilvum_PYR-GCK AAAWTRKGVRVNCIACGAVRTPGLEADAKRQGFDIDMIGQTNGSGRIAEP
TH_4266|M.thermoresistible__bu AAAWTRKGVRVNCIACGAIRTPGLEADAKRQGFDIDMIGQTNGSGRIAEP
MAB_4632|M.abscessus_ATCC_1997 AMDLSPR-IRVNGIAPGSILTSALEVVAGNDEMRN-TLEQNTPLHRLGDP
Mb1963c|M.bovis_AF2122/97 AVELAPHKIRVNSVSPGYILTELVEP---YTEYQP-LWEPKIPLGRLGRP
Rv1928c|M.tuberculosis_H37Rv AVELAPHKIRVNSVSPGYILTELVEP---YTEYQP-LWEPKIPLGRLGRP
MMAR_2849|M.marinum_M AVELAPHQIRVNSVSPGYIRTELVEP---LADYHA-LWEPKIPLGRMGRP
MAV_2775|M.avium_104 AVEFAPHDIRVNSVSPGYIRTELVEP---LHEYHR-QWQPKIPLGRMGRP
MSMEG_3604|M.smegmatis_MC2_155 AAEWAPLGIRVNALAPGYVKTDMAP--VDRPEFKR-YWIDDTPQLRYAVP
Mvan_0894|M.vanbaalenii_PYR-1 AVQWARYNVRVNALAPGFFPSELTTELLDDDAGGA-YIRRNTPMARAGRA
MLBr_01807|M.leprae_Br4923 ARELGSRSITANVVAPGFIDTEMTRAMDSKYQEAA---LQAIPLGRIGAV
MUL_3898|M.ulcerans_Agy99 AKELAHVGVRVNAIAPGLIRSAMTEAMPQRIWDQK---LAEVPMGRAGEP
* : .* :: * . : * .
Mflv_1263|M.gilvum_PYR-GCK DEIGYGVLFFASDASSYCSGQTLYMHGGPGPAGV--------
TH_4266|M.thermoresistible__bu DEIGYGVLFFASDASSYCSGQTLYMHGGPGPAGV--------
MAB_4632|M.abscessus_ATCC_1997 ADIAAAAVYLASPAGSFLTGKVLEVDGGLIAPNLDIPLPDLA
Mb1963c|M.bovis_AF2122/97 EELAGLYLYLASEASSYMTGSDIVIDGGYTCP----------
Rv1928c|M.tuberculosis_H37Rv EELAGLYLYLASEASSYMTGSDIVIDGGYTCP----------
MMAR_2849|M.marinum_M EELTGLYLYLASAASSYMTGSDIVIDGGYTCP----------
MAV_2775|M.avium_104 DELAGIYLYLASAASSYMTGSDIVIDGGYSCP----------
MSMEG_3604|M.smegmatis_MC2_155 EEIAPSVVFLASDAASFITGSVLVADGGYTAW----------
Mvan_0894|M.vanbaalenii_PYR-1 GELSGALLLLASGAGSFITGQVIAVDGGWTAR----------
MLBr_01807|M.leprae_Br4923 EEIAGAVSFLASEDAGYISGAVIPVDGGLGMGH---------
MUL_3898|M.ulcerans_Agy99 SEVASVALFLACDLSSYMTGTVLDVTGGRFI-----------
:: :*. ..: :* : **