For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MNYQDAIAGLDAKHGILIDGNARGADATFDVHDPATGEVIAAVSDGSAADAVAAVDAAQKAFGAWAATSP RERSEILRRAYELILADIDRLSALVCAENGKSQADARAEVTYAAEFFRWFSEEAVRTDGAFGSNPAGNAR TLVTHKPVGVAALVTPWNFPAAMATRKIAPALAAGCTVVLKPAAETPLTALAIAAILTDAGVPAGVVNLV PTSDAGAVVSAWLADARVRKISFTGSTGIGRVLLKQAADRVVNASMELGGNAPFVVTADADIAKAVEGAM LAKFRGAGQVCTAANRFYVHASVHDEFVAAFAAKIEALRVGPAADPASEIGPLVSPRAAQRVSAAIDAAV ADGAVVAAKAPTPTEGWFVAPTLLTDVAPDAAILTDEIFGPVAPVVAWEDEDELLRWANDTEYGLAAYVF AGRLQDALRIAERIDAGMVGVNRGAVSDPSAPFGGMKQSGLGREGAREGLQEFQETQYFSVAFD
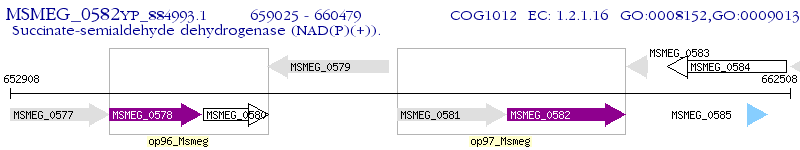
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0582 | - | - | 100% (484) | succinic semialdehyde dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5538 | - | e-135 | 49.59% (482) | [NADP+] succinate-semialdehyde dehydrogenase [Mycobacterium |
| M. smegmatis MC2 155 | MSMEG_2488 | - | e-132 | 51.39% (469) | [NADP+] succinate-semialdehyde dehydrogenase [Mycobacterium |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0791 | aldA | 2e-74 | 36.98% (484) | aldehyde dehydrogenase NAD dependant AldA |
| M. gilvum PYR-GCK | Mflv_1858 | - | 1e-136 | 50.85% (468) | succinate-semialdehyde dehydrogenase (NAD(P)(+)) |
| M. tuberculosis H37Rv | Rv0223c | - | 3e-77 | 36.24% (458) | aldehyde dehydrogenase |
| M. leprae Br4923 | MLBr_02573 | gabD1 | 1e-72 | 35.14% (444) | succinic semialdehyde dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4208 | - | 1e-135 | 50.66% (454) | putative succinate-semialdehyde dehydrogenase |
| M. marinum M | MMAR_0472 | - | 1e-76 | 36.50% (463) | aldehyde dehydrogenase |
| M. avium 104 | MAV_2101 | - | 3e-82 | 37.88% (462) | aldehyde dehydrogenase |
| M. thermoresistible (build 8) | TH_4137 | - | 1e-112 | 45.34% (472) | PUTATIVE putative [NADP+] succinate-semialdehyde dehydrogenase |
| M. ulcerans Agy99 | MUL_1122 | - | 8e-75 | 35.85% (463) | aldehyde dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_4876 | - | 1e-135 | 50.85% (468) | succinate-semialdehyde dehydrogenase (NAD(P)(+)) |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1858|M.gilvum_PYR-GCK -MDNSAVEALLASVPTGLWIGGEEREGTS--TFDVLDPSDDTVLTSVADA
Mvan_4876|M.vanbaalenii_PYR-1 -MDTS---ALLKSVPTGLWIGGEERQGTS--TFSVLDPSDDEVLATVADA
MAB_4208|M.abscessus_ATCC_1997 MTDHS---TLIDAIPTDLWIGGKQVPSSTGTRFPVHNPATGAVLTTVADA
MSMEG_0582|M.smegmatis_MC2_155 -MNYQDAIAGLDAK-HGILIDGNARGADA--TFDVHDPATGEVIAAVSDG
TH_4137|M.thermoresistible__bu -----------------MYIDGEWQSAASGKTFASINPANGAVLDSVPDG
MMAR_0472|M.marinum_M --------MGEKTEYDKLFIGGKWTEPSTSEVIEVHCPATGEYVGKVPLA
MUL_1122|M.ulcerans_Agy99 --------MGEKTEYDKLFIGGRWTEPSTSEVIEVHCPATGEYVGKVPLA
Rv0223c|M.tuberculosis_H37Rv -------MSDSATEYDKLFIGGKWTKPSTSDVIEVRCPATGEYVGKVPMA
MAV_2101|M.avium_104 ----------------------------MSTTIPVFNPSTEEQIAEVPDS
Mb0791|M.bovis_AF2122/97 -------MALWGDGISALLIDGKLSD-GRAGTFPTVNPATEEVLGVAADA
MLBr_02573|M.leprae_Br4923 ------------------------------MSIATINPATGETVKTFTPT
: *: : .
Mflv_1858|M.gilvum_PYR-GCK TADDARAALDAACAVQA--EWAGTAPRKRGEILRSVFETITARADDIAAL
Mvan_4876|M.vanbaalenii_PYR-1 TADDARDALDAACAVQG--EWAATPARKRGEILRSVFETITARADDIAAL
MAB_4208|M.abscessus_ATCC_1997 AASDGAIALDEATAVQR--SWAATAPRERAEILRRAWELVIARRDDFALA
MSMEG_0582|M.smegmatis_MC2_155 SAADAVAAVDAAQKAFG--AWAATSPRERSEILRRAYELILADIDRLSAL
TH_4137|M.thermoresistible__bu EREDARRAIDAAAAALP--DWRSRTAYERAEVLVEAHRIMLERRDDLAEL
MMAR_0472|M.marinum_M AAADVDAAVAAARAAFDNGPWPSTPPKERATVIANALKLLEERKDHFAKL
MUL_1122|M.ulcerans_Agy99 AAADVDAAVAAARAAFDNGPWPSTPPKERATVIANALKLLEERKDHFAKL
Rv0223c|M.tuberculosis_H37Rv AAADVDAAVAAARAAFDNGPWPSTPPHERAAVIAAAVKMLAERKDLFTKL
MAV_2101|M.avium_104 DQAAVDAAVARARETFESGVWRKAPASHRANVLFRAARIIEERKDELAAL
Mb0791|M.bovis_AF2122/97 DAEDMGRAIEAARRAFDSTDWSRN-TELRVRCVRQLRDAMQQHVEELREL
MLBr_02573|M.leprae_Br4923 TQDEVEDAIARAYARFDN--YRHTTFTQRAKWANATADLLEAEANQSAAL
*: * : * : :
Mflv_1858|M.gilvum_PYR-GCK MTLEMGKV-LAESKGEVTYGAEFFRWFAEEAVRIDG-----RYTAAPAG-
Mvan_4876|M.vanbaalenii_PYR-1 MTLEMGKV-FAESKGEVTYGAEFFRWFAEEAVRIEG-----RYTPAPAG-
MAB_4208|M.abscessus_ATCC_1997 MTLEMGKP-LAESQGEVAYGGEFLRWFSEEAVRING-----RYTSSPTG-
MSMEG_0582|M.smegmatis_MC2_155 VCAENGKS-QADARAEVTYAAEFFRWFSEEAVRTDG-----AFGSNPAG-
TH_4137|M.thermoresistible__bu MTREQGKP-LRMARTEVGYAADFLLWFAEEAKRVYG-----EVIPSARA-
MMAR_0472|M.marinum_M LADETGQPPMTVETMHWMGSIGALNFFAGPAVDQVR---WKEIRNGSYG-
MUL_1122|M.ulcerans_Agy99 FADETGQPPMTVETMHWMGSIGALNFFAGPAVDQVR---WKEIRNGSYG-
Rv0223c|M.tuberculosis_H37Rv LAAETGQPPTIIETMHWMGSMGAMNYFAG-AADKVT---WTETRTGSYG-
MAV_2101|M.avium_104 ESQDNGMN-LTAAGHIIPVSVDMLKYYAG-WVGKIHGESANLVSDGLLGT
Mb0791|M.bovis_AF2122/97 TISEVGAPRMLTASAQLEGPVGDLSFAADTAESYPWK--QDLGEASPLG-
MLBr_02573|M.leprae_Br4923 MTLEMGKT-LQSAKDEAMKCAKGFRYYAEKAETLLAD---EPAEAAAVG-
: * : : : .
Mflv_1858|M.gilvum_PYR-GCK --TGRILVTKQAVGPCYAITPWNFPLAMGTRKMGPAFAAGCTMIVKPAQE
Mvan_4876|M.vanbaalenii_PYR-1 --TGRILVTKQAVGPCYAITPWNFPLAMGTRKMGPAFAAGCTMIVKPAQE
MAB_4208|M.abscessus_ATCC_1997 --AGRILVAKQPIGPTLAITPWNFPLAMGTRKIGPALAAGCTMIVKPAAE
MSMEG_0582|M.smegmatis_MC2_155 --NARTLVTHKPVGVAALVTPWNFPAAMATRKIAPALAAGCTVVLKPAAE
TH_4137|M.thermoresistible__bu --DQRFIVLAQPVGVCAAITPWNYPISMLTRKLAPALAAGCTSVLKPAEQ
MMAR_0472|M.marinum_M ----QTIVHREPIGVVGAIVAWNVPLFLAINKLGPALLAGCTVVLKPAAE
MUL_1122|M.ulcerans_Agy99 ----QTIVHREPIGVVGAIVAWNVPLFLAINKLGPAPLAGCTVVLKPAAE
Rv0223c|M.tuberculosis_H37Rv ----QSIVSREPVGVVGAIVAWNVPLFLAVNKIAPALLAGCTIVLKPAAE
MAV_2101|M.avium_104 WETYHTFTQLEPVGVVGLIIPWNGPFFVAMLKVAPALAAGCSAVLKPAEE
Mb0791|M.bovis_AF2122/97 -IATRRTLAREAVGVVGAITPWNFPHQINLAKLGPALAAGNTVVLKPAPD
MLBr_02573|M.leprae_Br4923 --ASKALARYQPLGVVLAVMPWNFPLWQTVRFAAPALMAGNVGLLKHASN
: :.:* : .** * .** ** ::* * :
Mflv_1858|M.gilvum_PYR-GCK TPLTMLLLAKLMADAG-LPKGVLSVLPTTSPGPVTETLIDDGRLRKLTFT
Mvan_4876|M.vanbaalenii_PYR-1 TPLTMLLLAKLMAEAG-LPKGVLSVLPTSNPGAVTEALINDGRLRKLTFT
MAB_4208|M.abscessus_ATCC_1997 TPLTMLLLGQVFADAG-LPAGVLSILPTTNASALTGPLIDDPRLRKLTFT
MSMEG_0582|M.smegmatis_MC2_155 TPLTALAIAAILTDAG-VPAGVVNLVPTSDAGAVVSAWLADARVRKISFT
TH_4137|M.thermoresistible__bu TPLCAVETFKVFADAG-VPAGVVNLVTCSDPEPVADELLGDPRVRKISFT
MMAR_0472|M.marinum_M TPLTANALAEVFAEAG-LPEGVLSVVPG--GVETGQALTSNPGVDLFTFT
MUL_1122|M.ulcerans_Agy99 TPLTANALAEVFAEAG-LPEGVLSVVPG--GVETGQALTSNPGVDLFTFT
Rv0223c|M.tuberculosis_H37Rv TPLTANALAEVFAEVG-LPEGVLSVVPG--GIETGQALTSNPDIDMFTFT
MAV_2101|M.avium_104 TPLTALKLEEIFREAG-LPDGVLNVITGY-GETTGAALTAHPDVDKISFT
Mb0791|M.bovis_AF2122/97 TPWCAAALGEIIVEHTDFPPGVVNIVTSS-SHALGALLAKDPRVDMISFT
MLBr_02573|M.leprae_Br4923 VPQCALYLADVIARGG-FPEDCFQTLLIS--ANGVEAILRDPRVAAATLT
.* :: .* . .. : . : ::*
Mflv_1858|M.gilvum_PYR-GCK GSTGVGKALVKQSADKLLRTSMELGGNAPFVVFDDADVDAAVDGAILAKM
Mvan_4876|M.vanbaalenii_PYR-1 GSTGVGKALVKQSADKLLRTSMELGGNAPFIVFDDADVDAAVDGAILAKM
MAB_4208|M.abscessus_ATCC_1997 GSTGVGKILLSQCATRVLRSSMELGGNAPFVVFDDADVDAAVDGAMAAKM
MSMEG_0582|M.smegmatis_MC2_155 GSTGIGRVLLKQAADRVVNASMELGGNAPFVVTADADIAKAVEGAMLAKF
TH_4137|M.thermoresistible__bu GSTEVGRMIASRAGATMKRVSMELGGHAPFLVFPDADPEHAAKGGAAVKF
MMAR_0472|M.marinum_M GSSAVGKEIGRRAAEMLKPCTLELGGKSAAILLEDVDLSTAVPMMVFSGI
MUL_1122|M.ulcerans_Agy99 GSSAVGKEIGRRAAEMLKPCTLELGGKSAAILLEDVDLSTAVPMMVFSGI
Rv0223c|M.tuberculosis_H37Rv GSSAVGREVGRRAAEMLKPCTLELGGKSAAIILEDVDLAAAIPMMVFSGV
MAV_2101|M.avium_104 GSTEVGRLIVKAAAGNLKRLTLELGGKSPLIMFDDANLDKAIMGAGMGLL
Mb0791|M.bovis_AF2122/97 GSTATGRAVMADAAATIKKVFLELGGKSAFVVLDDADLAAASAVSAFSAC
MLBr_02573|M.leprae_Br4923 GSEPAGQAVGAIAGNEIKPTVLELGGSDPFIVMPSADLDAAVATAVTGRV
** *: : .. : :**** . :: ..: *
Mflv_1858|M.gilvum_PYR-GCK RNGGEACTAANRIHVANAVREEFTDKFVKRMSEFTLGKGLDEKSTLGPLI
Mvan_4876|M.vanbaalenii_PYR-1 RNGGEACTAANRIHVANAVREEFTEKFVKRMSEFTLGKGLDEKSTLGPLI
MAB_4208|M.abscessus_ATCC_1997 RNIGEACTAANRFHVDNAVRAEFTEKLTARMGALTIGPGDENGVQVGPLI
MSMEG_0582|M.smegmatis_MC2_155 RGAGQVCTAANRFYVHASVHDEFVAAFAAKIEALRVGPAADPASEIGPLV
TH_4137|M.thermoresistible__bu LNTGQACICPNRFIVHRSIAEPFIATLRARIEKLVPGDGLAAGTTVGPLI
MMAR_0472|M.marinum_M MNTGQACVGQTRILAPRSRYEEIVEAVSAFVQALPVGPPSDPAAQIGSLI
MUL_1122|M.ulcerans_Agy99 MNTGQACVGQTRILAPRSRYEEIVEAVSAFVQTLPVGPPSDPAAQIGSLI
Rv0223c|M.tuberculosis_H37Rv MNAGQGCVNQTRILAPRSRYDEIVAAVTNFVTALPVGPPSDPAAQIGPLI
MAV_2101|M.avium_104 AGSGQNCSCTSRIYVQRGIYDQVVEGLAEFAMMLPMGGSDDPNSVLGPLI
Mb0791|M.bovis_AF2122/97 MHAGQGCAITTRLVVPRARYEEAVAIAAATMSSIRPGDPNDPGTVCGPLI
MLBr_02573|M.leprae_Br4923 QNNGQSCIAAKRFIAHADIYDEFVEKFVARMETLRVGDPTDPDTDVGPLA
*: * .*: . : * *.*
Mflv_1858|M.gilvum_PYR-GCK NAKQVATVTELVSDAVSRGATVAVGG----VAPGGPGNFYPATVLTDVPA
Mvan_4876|M.vanbaalenii_PYR-1 NAKQVATVTELVSDAVSRGATVAVGG----VAPGGPGNFYPATVLTDVPA
MAB_4208|M.abscessus_ATCC_1997 TAKQRASVDELVQDAVSKGAVVTTGG----VVADGEGYFYPPTVLTDVPA
MSMEG_0582|M.smegmatis_MC2_155 SPRAAQRVSAAIDAAVADGAVVAAKA----PTP-TEGWFVAPTLLTDVAP
TH_4137|M.thermoresistible__bu DEVALKKMQRQVDDAVDKGAKLELGGSRLQGPAFDNGHFYAPTILTGVTP
MMAR_0472|M.marinum_M SEKQRARVEGYIAKGIEEGARLVSGGG--RPEGLDNGFFVQPTVFADVDN
MUL_1122|M.ulcerans_Agy99 SEKQRARVEGYIAKGIEEGARLVSGGG--RPESLDNGFFVQPTVFADVDN
Rv0223c|M.tuberculosis_H37Rv SEKQRTRVEGYIAKGIEEGARLVCGGG--RPEGLDNGFFIQPTVFADVDN
MAV_2101|M.avium_104 SEKQRQRVEGIVNDGVAAGAKVITGG---KPMDRK-GYFYEATIITNTTP
Mb0791|M.bovis_AF2122/97 SARQRDRVQGYLDLAVAEGGRFACGGA--RPADREVGFYIEPTVIAGLTN
MLBr_02573|M.leprae_Br4923 TESGRAQVEQQVDAAAAAGAVIRCGGK----RPDRPGWFYPPTVITDITK
: : . *. . . * : .*:::.
Mflv_1858|M.gilvum_PYR-GCK DARILKEEVFGPVAPIAGFDTEEEGIAAANDTEYGLAAYVYTQSLDRALR
Mvan_4876|M.vanbaalenii_PYR-1 DARILKEEVFGPVAPIAGFDTEEEGIAAANDTEYGLAAYVYTQSLDRALR
MAB_4208|M.abscessus_ATCC_1997 DARILREEVFGPVAAITGFDGEEAGIAAANDTEFGLAAYVYTANLDRALR
MSMEG_0582|M.smegmatis_MC2_155 DAAILTDEIFGPVAPVVAWEDEDELLRWANDTEYGLAAYVFAGRLQDALR
TH_4137|M.thermoresistible__bu DMLICSEETFGPIAPVMVFDDEDEAIAMANSTEYGLAGYIYTRDISRAIR
MMAR_0472|M.marinum_M KMTIAQEEIFGPVLVVIPYDTEEDAIKIANDSVYGLAGSVWTSNVPKGIE
MUL_1122|M.ulcerans_Agy99 KMTIAQEEIFGPVLVVIPYDTKEDAIKIANDSVYGLAGSVWTSNVPKGIE
Rv0223c|M.tuberculosis_H37Rv KMTIAQEEIFGPVLAIIPYDTEEDAIAIANDSVYGLAGSVWTTDVPKGIK
MAV_2101|M.avium_104 DMRLIREEIFGPVGCVIPFDDEDEVIAAANDTHYGLAGAIWTENLSRAHR
Mb0791|M.bovis_AF2122/97 DARVAREEIFGPVLTVIAHDGDDDAVRIANDSPYGLSGTVYGADPQRAAR
MLBr_02573|M.leprae_Br4923 DMALYTDEVFGPVASVYCATNIDDAIEIANATPFGLGSNAWTQNESEQRH
. : :* ***: : : : ** : :**.. : .
Mflv_1858|M.gilvum_PYR-GCK VAEGIESGMVGVNRGVISDPAAPFGGIKESGFGREGGTEGIEEYLDTKYI
Mvan_4876|M.vanbaalenii_PYR-1 VAENIESGMVGVNRGVISDAAAPFGGIKESGFGREGGTEGIEEYLDTKYI
MAB_4208|M.abscessus_ATCC_1997 VSQAVESGMVGVNRGVISDVAAPFGGIKESGIGREAGSEGIEEYLETKYI
MSMEG_0582|M.smegmatis_MC2_155 IAERIDAGMVGVNRGAVSDPSAPFGGMKQSGLGREGAREGLQEFQETQYF
TH_4137|M.thermoresistible__bu VGEALDFGMIGINDINPTSAAVPFGGIKQSGLGREGARAGIEEYLDTKVL
MMAR_0472|M.marinum_M ISEKIRTGTYAIN-WYAFDPCCPFGGYKNSGIGRENGPEGVEHFTQQKSV
MUL_1122|M.ulcerans_Agy99 ISEKIRTGTYAIN-WYAFDPCCPFGGYKNSGIGRENGPEGVEHFTQQKSV
Rv0223c|M.tuberculosis_H37Rv ISQQIRTGTYGIN-WYAFDPGSPFGGYKNSGIGRENGPEGVEHFTQQKSV
MAV_2101|M.avium_104 VVNELRAGQIWVNSALAADPSMPISGHKQSGWGGERGRKGIEAYFNTKAV
Mb0791|M.bovis_AF2122/97 IASRLRVGTVNVNGGVWYCADAPFGGYKQSGIGREMGLLGFEEYLEAKLI
MLBr_02573|M.leprae_Br4923 FIDNIVAGQVFINGMTVSYPELPFGGIKRSGYGRELSTHGIREFCNIKTV
. . : * :* *:.* *.** * * . *.. : : : .
Mflv_1858|M.gilvum_PYR-GCK ALTK------
Mvan_4876|M.vanbaalenii_PYR-1 ALTR------
MAB_4208|M.abscessus_ATCC_1997 ALP-------
MSMEG_0582|M.smegmatis_MC2_155 SVAFD-----
TH_4137|M.thermoresistible__bu GIAL------
MMAR_0472|M.marinum_M LMPMGYTLES
MUL_1122|M.ulcerans_Agy99 LMPMGYTLES
Rv0223c|M.tuberculosis_H37Rv LLPMGYTVA-
MAV_2101|M.avium_104 YIGL------
Mb0791|M.bovis_AF2122/97 ATAAN-----
MLBr_02573|M.leprae_Br4923 WIA-------