For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GEKTEYDKLFIGGRWTEPSTSEVIEVHCPATGEYVGKVPLAAAADVDAAVAAARAAFDNGPWPSTPPKER ATVIANALKLLEERKDHFAKLFADETGQPPMTVETMHWMGSIGALNFFAGPAVDQVRWKEIRNGSYGQTI VHREPIGVVGAIVAWNVPLFLAINKLGPAPLAGCTVVLKPAAETPLTANALAEVFAEAGLPEGVLSVVPG GVETGQALTSNPGVDLFTFTGSSAVGKEIGRRAAEMLKPCTLELGGKSAAILLEDVDLSTAVPMMVFSGI MNTGQACVGQTRILAPRSRYEEIVEAVSAFVQTLPVGPPSDPAAQIGSLISEKQRARVEGYIAKGIEEGA RLVSGGGRPESLDNGFFVQPTVFADVDNKMTIAQEEIFGPVLVVIPYDTKEDAIKIANDSVYGLAGSVWT SNVPKGIEISEKIRTGTYAINWYAFDPCCPFGGYKNSGIGRENGPEGVEHFTQQKSVLMPMGYTLES*
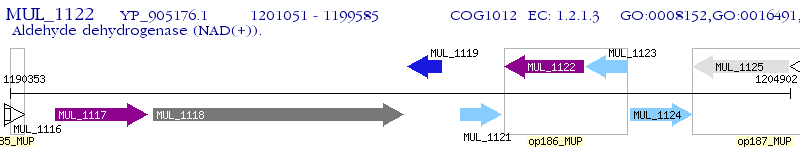
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_1122 | - | - | 100% (488) | aldehyde dehydrogenase |
| M. ulcerans Agy99 | MUL_0939 | aldA_1 | 1e-88 | 39.46% (479) | NAD-dependent aldehyde dehydrogenase, AldA_1 |
| M. ulcerans Agy99 | MUL_2877 | - | 2e-88 | 41.01% (456) | aldehyde dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0228c | - | 1e-162 | 79.21% (356) | aldehyde dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0428 | - | 0.0 | 69.09% (482) | betaine-aldehyde dehydrogenase |
| M. tuberculosis H37Rv | Rv0223c | - | 0.0 | 84.23% (482) | aldehyde dehydrogenase |
| M. leprae Br4923 | MLBr_02573 | gabD1 | 7e-60 | 33.19% (452) | succinic semialdehyde dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4484 | - | 0.0 | 70.25% (484) | aldehyde dehydrogenase |
| M. marinum M | MMAR_0472 | - | 0.0 | 98.77% (488) | aldehyde dehydrogenase |
| M. avium 104 | MAV_4946 | - | 0.0 | 88.84% (484) | aldehyde dehydrogenase family protein |
| M. smegmatis MC2 155 | MSMEG_0309 | - | 0.0 | 69.98% (483) | aldehyde dehydrogenase family protein |
| M. thermoresistible (build 8) | TH_4171 | - | 1e-131 | 76.79% (293) | PUTATIVE aldehyde dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0240 | - | 0.0 | 68.88% (482) | betaine-aldehyde dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_1122|M.ulcerans_Agy99 -----------MGEKTEYDKLFIGGRWTEPSTSEVIEVHCPATGEYVGKV
MMAR_0472|M.marinum_M -----------MGEKTEYDKLFIGGKWTEPSTSEVIEVHCPATGEYVGKV
MAV_4946|M.avium_104 ------MSDIQTQAKTEYDKLFIGGKWTEPSTSEVIEVHCPATGEYVGKV
Mb0228c|M.bovis_AF2122/97 ----------MSDSATEYDKLFIGGKWTKPSTSDVIEVRCPATGEYVGKV
Rv0223c|M.tuberculosis_H37Rv ----------MSDSATEYDKLFIGGKWTKPSTSDVIEVRCPATGEYVGKV
Mflv_0428|M.gilvum_PYR-GCK ---MTQS----TAFRTEWDKLFIGGKWVEPASSEVIEVRSPATGDVVGKV
Mvan_0240|M.vanbaalenii_PYR-1 ---MTQS----TTFRTEWDKLFIGGKWVEPATSDVIEVHSPATGELVGKV
MSMEG_0309|M.smegmatis_MC2_155 MTAMTQTAPAATAATTNWDKLFIGGQWVEPSSSEIIEVFSPATGEKVGQA
TH_4171|M.thermoresistible__bu --------------------------------------------------
MAB_4484|M.abscessus_ATCC_1997 --------------MTNYDKLFIGGVWTEPATDQVIEVISPATGQKVGQC
MLBr_02573|M.leprae_Br4923 ---------------------------------MSIATINPATGETVKTF
MUL_1122|M.ulcerans_Agy99 PLAAAADVDAAVAAARAAFDNGPWPSTPPKERATVIANALKLLEERKDHF
MMAR_0472|M.marinum_M PLAAAADVDAAVAAARAAFDNGPWPSTPPKERATVIANALKLLEERKDHF
MAV_4946|M.avium_104 PLAAAADVDAAVAAARAAFDNGPWPTTPPKERAAVIANALKVMEERKDLF
Mb0228c|M.bovis_AF2122/97 PMAAAADVDAAVAAARAAFDNGPWPSTPPHERAAVIAAAVKMLAERKDLF
Rv0223c|M.tuberculosis_H37Rv PMAAAADVDAAVAAARAAFDNGPWPSTPPHERAAVIAAAVKMLAERKDLF
Mflv_0428|M.gilvum_PYR-GCK PLASAADVDAACAAAREAFDNGPWPQMSPTERAEVLGRAVKLMEERADEL
Mvan_0240|M.vanbaalenii_PYR-1 PLASAADVDAACAAAREAFDNGPWPHMSPAERGEILGRAVKIMEERADEL
MSMEG_0309|M.smegmatis_MC2_155 PLATKADVDAACAAARKAFDEGPWPRMTPQEREAVLAKATALIEESADEF
TH_4171|M.thermoresistible__bu --------------------------------------------------
MAB_4484|M.abscessus_ATCC_1997 PLASPADVDAAVSAARRAFDNGPWPRLTPHERQAVLQKAVSLLESRKDEI
MLBr_02573|M.leprae_Br4923 TPTTQDEVEDAIARAYARFDN--YRHTTFTQRAKWANATADLLEAEANQS
MUL_1122|M.ulcerans_Agy99 AKLFADETGQPPMTVETMHWMGSIGALNFFAGPAVDQVRWKEIRNGSYG-
MMAR_0472|M.marinum_M AKLLADETGQPPMTVETMHWMGSIGALNFFAGPAVDQVRWKEIRNGSYG-
MAV_4946|M.avium_104 TGLLAAETGQPPTSVETMHWMSSIGALNFFAGPAVDQVKWKEVRTGGYG-
Mb0228c|M.bovis_AF2122/97 TKLLAAETGQPPTIIETMHWMGSMGAMNYFAG-AADKVTWTETRTGSYG-
Rv0223c|M.tuberculosis_H37Rv TKLLAAETGQPPTIIETMHWMGSMGAMNYFAG-AADKVTWTETRTGSYG-
Mflv_0428|M.gilvum_PYR-GCK KFLLAAETGQPPTIVDMMQYGAAMSSFQFYAG-AADKFTWQDIRDGVYG-
Mvan_0240|M.vanbaalenii_PYR-1 KFLLAAETGQPPTIVDMMQYGAAMSSFQFYAG-AADKFTWQDIRDGVYG-
MSMEG_0309|M.smegmatis_MC2_155 KTLLKLETGQPQTIVDMMQYGAAMSTLQFYAS-AADKFAWKDIRDGIYG-
TH_4171|M.thermoresistible__bu --------------------------------------------------
MAB_4484|M.abscessus_ATCC_1997 CKAIADETGAPPMVIETLQWMGGFGAIQYYAN-AADAVKWKELRNGAYG-
MLBr_02573|M.leprae_Br4923 AALMTLEMGKTLQSAKDEAMKCAKG-FRYYAEKAETLLADEPAEAAAVGA
MUL_1122|M.ulcerans_Agy99 -QTIVHREPIGVVGAIVAWNVPLFLAINKLGPAPLAGCTVVLKPAAETPL
MMAR_0472|M.marinum_M -QTIVHREPIGVVGAIVAWNVPLFLAINKLGPALLAGCTVVLKPAAETPL
MAV_4946|M.avium_104 -QSIVHREPIGVVGAIVAWNVPLFLAVNKLGPALLAGCTVVLKPAAETPL
Mb0228c|M.bovis_AF2122/97 -QSIVSRESVGVVGAIVAWNVPLFLAVNKIAPALLAGCTIVLKPAAETPL
Rv0223c|M.tuberculosis_H37Rv -QSIVSREPVGVVGAIVAWNVPLFLAVNKIAPALLAGCTIVLKPAAETPL
Mflv_0428|M.gilvum_PYR-GCK -QTLVVREPVGVVGAVTAWNVPFFLAANKLGPALLAGCTVVLKPAAETPL
Mvan_0240|M.vanbaalenii_PYR-1 -QTLVVREPVGVVGAVTAWNVPFFLAANKLGPALLAGCTVVLKPAAETPL
MSMEG_0309|M.smegmatis_MC2_155 -QTLVLKEPVGVVGAVVAWNVPFFLAANKLGPALLAGCTIVLKPAAETPL
TH_4171|M.thermoresistible__bu --------------------------------------------------
MAB_4484|M.abscessus_ATCC_1997 -QTLVTREPAGVVAAIAAWNVPLFLALNKIGPAFLAGCAVVLKPAAETPL
MLBr_02573|M.leprae_Br4923 SKALARYQPLGVVLAVMPWNFPLWQTVRFAAPALMAGNVGLLKHASNVPQ
MUL_1122|M.ulcerans_Agy99 TANALAEVFAEAGLPEGVLSVVPGGVETGQALTSNPGVDLFTFTGSSAVG
MMAR_0472|M.marinum_M TANALAEVFAEAGLPEGVLSVVPGGVETGQALTSNPGVDLFTFTGSSAVG
MAV_4946|M.avium_104 SANALAEAFAEAGLPEGVLSVVPGGIETGQALTSNTDVDIFSFTGSSAVG
Mb0228c|M.bovis_AF2122/97 TANALAEVFAEVGLPEGVLSVVPGGIETGQALTSNPDIDMFTFTGSSAVG
Rv0223c|M.tuberculosis_H37Rv TANALAEVFAEVGLPEGVLSVVPGGIETGQALTSNPDIDMFTFTGSSAVG
Mflv_0428|M.gilvum_PYR-GCK SVFAMAEMFVEAGLPEGVLSIVPGGPETGQALTANPNLDKYTFTGSSGVG
Mvan_0240|M.vanbaalenii_PYR-1 SVFAMAEMFVEAGLPEGVLSIVPGGPETGQALTANPNLDKYTFTGSSGVG
MSMEG_0309|M.smegmatis_MC2_155 TTNLMAQKFLEAGLPEGVLSVVPGGPETGRALTDNPALDKFTFTGSSAVG
TH_4171|M.thermoresistible__bu -------VFAEAGLPEGVLSVVPGGAETGRALTSNKDLDKYTFTGSSAVG
MAB_4484|M.abscessus_ATCC_1997 SSFILAEVFAEAGVPEGVLSVVPGGAETGRALTNNPDVDVFSFTGSTAVG
MLBr_02573|M.leprae_Br4923 CALYLADVIARGGFPEDCFQTLLISANGVEAILRDPRVAAATLTGSEPAG
: . *.**. :. : . : .*: : : ::*** .*
MUL_1122|M.ulcerans_Agy99 KEIGRRAAEMLKPCTLELGGKSAAILLEDVDLSTAVPMMVFSGIMNTGQA
MMAR_0472|M.marinum_M KEIGRRAAEMLKPCTLELGGKSAAILLEDVDLSTAVPMMVFSGIMNTGQA
MAV_4946|M.avium_104 REIGRRAADMLKPCTLELGGKSAAIVLDDVDLPSALPMLVFSGIMNSGQA
Mb0228c|M.bovis_AF2122/97 REVGRRAAEMLKPCTLELGGKSAAIILEDVDLAAAIPMMVFSGVMNAGQG
Rv0223c|M.tuberculosis_H37Rv REVGRRAAEMLKPCTLELGGKSAAIILEDVDLAAAIPMMVFSGVMNAGQG
Mflv_0428|M.gilvum_PYR-GCK KEIAKIAADKLKPCTLELGGKSAAIILEDADLDSTLPMLVFSGLMNSGQA
Mvan_0240|M.vanbaalenii_PYR-1 KEIAKIAADKLKPCTLELGGKSAAIILEDADLDSTLPMLVFSGLMNSGQA
MSMEG_0309|M.smegmatis_MC2_155 KEIGKIAAEKLKPCTLELGGKSAAIILEDADLDSTLPMLLFSGLMNSGQA
TH_4171|M.thermoresistible__bu REIGKIAAENLKPCTLELGGKSAAIILEDADLDATLPMLVFSGVMNSGQA
MAB_4484|M.abscessus_ATCC_1997 KEIAEIAAKNLKKVTLELGGKSAAIVLEDADLAAALPMLVFSGLMNAGQG
MLBr_02573|M.leprae_Br4923 QAVGAIAGNEIKPTVLELGGSDPFIVMPSADLDAAVATAVTGRVQNNGQS
: :. *.. :* .*****... *:: ..** :::. : . : * **.
MUL_1122|M.ulcerans_Agy99 CVGQTRILAPRSRYEEIVEAVSAFVQTLPVGPPSDPAAQIGSLISEKQRA
MMAR_0472|M.marinum_M CVGQTRILAPRSRYEEIVEAVSAFVQALPVGPPSDPAAQIGSLISEKQRA
MAV_4946|M.avium_104 CVAQTRILAPRSRYDEIVDAVSTFVQSLPVGLPSDPAAQIGSLISEKQRA
Mb0228c|M.bovis_AF2122/97 CVNQTRILAPRSRYDEIVAAVTNFVTALPVGPPSDPAAQIGPLISEKQRT
Rv0223c|M.tuberculosis_H37Rv CVNQTRILAPRSRYDEIVAAVTNFVTALPVGPPSDPAAQIGPLISEKQRT
Mflv_0428|M.gilvum_PYR-GCK CVGQTRILAPRSRYDEVIEKLGEAVRNMAPGLPDNPAAMIGPLISEKQRD
Mvan_0240|M.vanbaalenii_PYR-1 CVGQTRILAPRSRYDEVVEKLAAAAAGMAPGLPDNPAAMIGPLISEKQRD
MSMEG_0309|M.smegmatis_MC2_155 CVGQTRILAPRSRYDEVVEKVAAGVAAMQVGVPDDPAAMVGPLISEKQRE
TH_4171|M.thermoresistible__bu CVGQTRILAPRSRYDEVVEKVANFVAAMPVGLPDDPNTAIGPLISEKQRE
MAB_4484|M.abscessus_ATCC_1997 CVNQTRVLAPRSRYDEVVDGIVGMVSLMGVGLPDDPATQVGPLITEKQRT
MLBr_02573|M.leprae_Br4923 CIAAKRFIAHADIYDEFVEKFVARMETLRVGDPTDPDTDVGPLATESGRA
*: .*.:* . *:*.: . : * * :* : :*.* :*. *
MUL_1122|M.ulcerans_Agy99 RVEGYIAKGIEEGARLVSGGGRPESLDNGFFVQPTVFADVDNKMTIAQEE
MMAR_0472|M.marinum_M RVEGYIAKGIEEGARLVSGGGRPEGLDNGFFVQPTVFADVDNKMTIAQEE
MAV_4946|M.avium_104 RVEGYIAKGIEEGARLVCGGGRPEGLDSGFFVQPTVFADVDNKMTIAQEE
Mb0228c|M.bovis_AF2122/97 RVEALHRQG-HRGGRSVGVRRRP---------------------------
Rv0223c|M.tuberculosis_H37Rv RVEGYIAKGIEEGARLVCGGGRPEGLDNGFFIQPTVFADVDNKMTIAQEE
Mflv_0428|M.gilvum_PYR-GCK RVEGYIKKGIEEGARVITGGGRPEGLDSGWFVEPTVFADVDNSMTIAQEE
Mvan_0240|M.vanbaalenii_PYR-1 RVEGYIKKGIEEGARVVTGGGRPEGLDSGWYVQPTVFADVDNSMTIAQEE
MSMEG_0309|M.smegmatis_MC2_155 RVEGYIKKGIEEGARLVTGGGRPEGLDSGWFVQPTVFADVDNSMTIAQEE
TH_4171|M.thermoresistible__bu RVEGYIKKGIEEGARLVTGGGRPEGLDSGWFVQPTVFADVDNSMTIAQEE
MAB_4484|M.abscessus_ATCC_1997 RVEGYIAKGIAEGARLAYGGKRPEGLDGGYFVQPTIFADVDNSMTIAQEE
MLBr_02573|M.leprae_Br4923 QVEQQVDAAAAAGAVIRCGGKRPD--RPGWFYPPTVITDITKDMALYTDE
:** . *. **
MUL_1122|M.ulcerans_Agy99 IFGPVLVVIPYDTKEDAIKIANDSVYGLAGSVWTSNVPKGIEISEKIRTG
MMAR_0472|M.marinum_M IFGPVLVVIPYDTEEDAIKIANDSVYGLAGSVWTSNVPKGIEISEKIRTG
MAV_4946|M.avium_104 IFGPVLSIIPYDTEEDAIKIANDSVYGLAGSVWTADVQRGIAVSEKIRTG
Mb0228c|M.bovis_AF2122/97 ----------------SRGLGQRLLYPIHERRWR----------------
Rv0223c|M.tuberculosis_H37Rv IFGPVLAIIPYDTEEDAIAIANDSVYGLAGSVWTTDVPKGIKISQQIRTG
Mflv_0428|M.gilvum_PYR-GCK IFGPVLSVIPYEDEDDAVRIANDSVYGLAGSVYTTDNDRALKIARRIRTG
Mvan_0240|M.vanbaalenii_PYR-1 IFGPVLAVIPYEDEDDAVRIANDSVYGLAGSVYTTDNDRALKIARRIRTG
MSMEG_0309|M.smegmatis_MC2_155 IFGPVLVVIPFEDEDDAVRIANDSVYGLAGSVYTTDHAKGVEIASKVRTG
TH_4171|M.thermoresistible__bu IFGPVLCIIPYEDEDDAVRIANDSVYGLAGSVYTQDYPKALEIASKIRTG
MAB_4484|M.abscessus_ATCC_1997 IFGPVLSIIAYDSVDEAVKIANDSNYGLAGSVWTTDVQKGLEVAAQIRTG
MLBr_02573|M.leprae_Br4923 VFGPVASVYCATNIDDAIEIANATPFGLGSNAWTQNESEQRHFIDNIVAG
: :.: : : :
MUL_1122|M.ulcerans_Agy99 TYAIN-WYAFDPCCPFGGYKNSGIGRENGPEGVEHFTQQKSVLMPMGYTL
MMAR_0472|M.marinum_M TYAIN-WYAFDPCCPFGGYKNSGIGRENGPEGVEHFTQQKSVLMPMGYTL
MAV_4946|M.avium_104 TYAIN-WYAFDPCCPFGGYKNSGIGRENGPEGVEHFTQQKSVLMPMGYTL
Mb0228c|M.bovis_AF2122/97 ------------------------GKWHGQCG------------------
Rv0223c|M.tuberculosis_H37Rv TYGIN-WYAFDPGSPFGGYKNSGIGRENGPEGVEHFTQQKSVLLPMGYTV
Mflv_0428|M.gilvum_PYR-GCK TYAVN-MYAFDPCAPFGGYKNSGIGRENGWEGIEAYCEQKSILLPFGYTP
Mvan_0240|M.vanbaalenii_PYR-1 TYAVN-MYAFDPCAPFGGFKNSGIGRENGWEGIEAYCEPKSILLPFGYTP
MSMEG_0309|M.smegmatis_MC2_155 TYGIN-MYAFDPGAPFGGYKNSGIGRECGPEGIAGYCESKSVLLPFGYTP
TH_4171|M.thermoresistible__bu TYAVN-MYAFDPGAPFGGYKNSGIGRENGPEGIEAYCQQKSVLLPFGYTP
MAB_4484|M.abscessus_ATCC_1997 TYGIN-WYAFDPSCPFGGYKQSGIGRENGPEGIEEYCELKSVLLPMGYTL
MLBr_02573|M.leprae_Br4923 QVFINGMTVSYPELPFGGIKRSGYGRELSTHGIREFCNIKTVWIA-----
*: . *
MUL_1122|M.ulcerans_Agy99 ES-
MMAR_0472|M.marinum_M ES-
MAV_4946|M.avium_104 EG-
Mb0228c|M.bovis_AF2122/97 ---
Rv0223c|M.tuberculosis_H37Rv A--
Mflv_0428|M.gilvum_PYR-GCK PAS
Mvan_0240|M.vanbaalenii_PYR-1 AT-
MSMEG_0309|M.smegmatis_MC2_155 E--
TH_4171|M.thermoresistible__bu ED-
MAB_4484|M.abscessus_ATCC_1997 ESI
MLBr_02573|M.leprae_Br4923 ---