For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTSPVMDWDDAYQQQGAFEGPPPWNIGEPQPELAALIAAGKIHSDVLDAGCGYAELSLALAAEGYTVVGI DITPTAIAAATKAAAERGLGNTSFMQADITDFNAYPAGSAGRFATVIDSTLFHSLPVSGRDDYLKSVHRA AAPGASYFVLVFAKGAFPAALETKPNEVDEDELRAAVGKYWEIDEIRPAFIHANVPVIPPLPAGSPIEFP PHQRDEKGRMKFPAYLLTAHKA
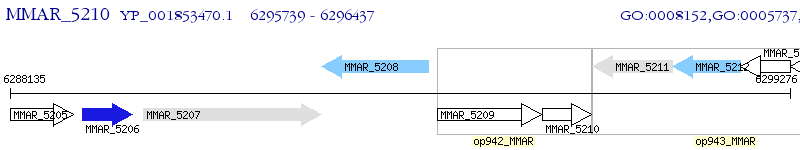
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_5210 | - | - | 100% (232) | hypothetical protein MMAR_5210 |
| M. marinum M | MMAR_0909 | - | 2e-61 | 55.90% (229) | methyltransferase |
| M. marinum M | MMAR_0089 | - | 6e-36 | 61.79% (123) | hypothetical protein MMAR_0089 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3725 | - | 1e-112 | 82.76% (232) | hypothetical protein Mb3725 |
| M. gilvum PYR-GCK | Mflv_1324 | - | 4e-77 | 63.16% (228) | methyltransferase type 11 |
| M. tuberculosis H37Rv | Rv3699 | - | 1e-112 | 82.76% (232) | hypothetical protein Rv3699 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0961c | - | 5e-74 | 62.39% (226) | hypothetical protein MAB_0961c |
| M. avium 104 | MAV_0404 | - | 2e-97 | 76.32% (228) | thiopurine S-methyltransferase (tpmt) superfamily protein |
| M. smegmatis MC2 155 | MSMEG_6235 | - | 3e-89 | 68.40% (231) | thiopurine S-methyltransferase (tpmt) superfamily protein |
| M. thermoresistible (build 8) | TH_1722 | - | 7e-83 | 66.52% (230) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_0662 | - | 7e-57 | 57.79% (199) | methyltransferase |
| M. vanbaalenii PYR-1 | Mvan_5471 | - | 8e-80 | 65.35% (228) | methyltransferase type 11 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1324|M.gilvum_PYR-GCK ---MA-------EVMDWDSAYRGEG-EFEG--EPPWNIGEPQPELAALWR
Mvan_5471|M.vanbaalenii_PYR-1 MAHMT-------DVMDWDSAYRGEG-DFEG--EPPWNIGEPQPELAALWR
Mb3725|M.bovis_AF2122/97 ---MT------DEVMDWDSAYREQG-AFEG--PPPWNIGEPQPELATLIA
Rv3699|M.tuberculosis_H37Rv ---MT------DEVMDWDSAYREQG-AFEG--PPPWNIGEPQPELATLIA
MMAR_5210|M.marinum_M ---MT------SPVMDWDDAYQQQG-AFEG--PPPWNIGEPQPELAALIA
MAV_0404|M.avium_104 --MSS------DQVMDWDSAYREQA-HFEG--PPPWNIGEPQPELAALIE
MSMEG_6235|M.smegmatis_MC2_155 MGVMT------SHVLDWDGAYRGDT-GFQG--PPPWNIGEPQPELAALHR
TH_1722|M.thermoresistible__bu MATEDPGTGDPTDTIDWDSVYRQEAPGFDG--PPPWNIGEPQPELAALHR
MAB_0961c|M.abscessus_ATCC_199 ---MTS-----NELPDWDATYQGKGELFQG--EPPWNIGEPQPELAALID
MUL_0662|M.ulcerans_Agy99 ---MT-----ESLDLQFATAYRGESAEFGAGIRPPWSLGEPQPELATLIE
:: .*: . * . ***.:********:*
Mflv_1324|M.gilvum_PYR-GCK DGRFRSEVLDAGCGHAELSLALAADGYTVTGVDISPTAMAAASAAAAERG
Mvan_5471|M.vanbaalenii_PYR-1 DGKFRSEVLDAGCGHAELSLALAADGYTVTGVDLSPTAVAAATAASAERG
Mb3725|M.bovis_AF2122/97 AGKVRSDVLDAGCGYAELSLALAADGYTVVGIDLTPTAVAAATKAAEERG
Rv3699|M.tuberculosis_H37Rv AGKVRSDVLDAGCGYAELSLALAADGYTVVGIDLTPTAVAAATKAAEERG
MMAR_5210|M.marinum_M AGKIHSDVLDAGCGYAELSLALAAEGYTVVGIDITPTAIAAATKAAAERG
MAV_0404|M.avium_104 QGKFRSDVLDAGCGCAELSLALAARGYTVVGIDLTPTAVAAATRAAAERG
MSMEG_6235|M.smegmatis_MC2_155 AGRFRSDVLDAGCGHAELSLALAADGYTVVGMDLSPSAIAAANRAAQERN
TH_1722|M.thermoresistible__bu AGAFRSDVLDAGCGHAEISMALAADGYTVVGIDLSPTAIAAANRTAQERG
MAB_0961c|M.abscessus_ATCC_199 AGKFHGTVLDVGCGHAETSLRLAALGHTTVGLDLSPTAIEAARAAAAERG
MUL_0662|M.ulcerans_Agy99 QGKVHGEVLDAGCGEAALAMALAGRGHPVVGLDISPTAVELAGREAARPG
* .:. ***.*** * :: **. *:...*:*::*:*: * : . .
Mflv_1324|M.gilvum_PYR-GCK LSEKASFVAADITALSGF----DGRFSTVVDSTLFHSLPVESRAGYLESV
Mvan_5471|M.vanbaalenii_PYR-1 LSERTSFVAADITTLTGF----DGRFATVVDSTLFHSLPVESRDGYLQSV
Mb3725|M.bovis_AF2122/97 LTT-ASFVQADITEFAAYPAGSAGRFSTVIDSTLFHSLPVDSRDRYLSSV
Rv3699|M.tuberculosis_H37Rv LTT-ASFVQADITEFAAYPAGSAGRFSTVIDSTLFHSLPVDSRDRYLSSV
MMAR_5210|M.marinum_M LGN-TSFMQADITDFNAYPAGSAGRFATVIDSTLFHSLPVSGRDDYLKSV
MAV_0404|M.avium_104 LTT-ASFVQADITSFTGY----DGRFATVVDSTLFHSLPVEGRDGYLRSI
MSMEG_6235|M.smegmatis_MC2_155 LAT-ASFVQGDITSFTGY----DGRFSTVIDSTLFHSLPVEGREGYLQSI
TH_1722|M.thermoresistible__bu LAT-ASFVQGDITDFGGY----DGRFNTIIDSTLFHSIPVQARDGYLRSI
MAB_0961c|M.abscessus_ATCC_199 LTN-ATFEVADITDFSGY----DGRFNTIVDSTLFHSIPVEAREAYLQSI
MUL_0662|M.ulcerans_Agy99 LTN-ASFAVADITDFASYPPESAGRFNTIMDSTLFHSMPVELREGCQRSI
* ::* .*** : .: *** *::*******:**. * *:
Mflv_1324|M.gilvum_PYR-GCK HRAARPGAGFFVLVFAKGAFPADTEQGP--NEVSEIELREAVSPYWTIDD
Mvan_5471|M.vanbaalenii_PYR-1 HRAAAPGAGYFVLVFAKGAFPAEMEGGP--NEVTELELREAVSRYWIIDD
Mb3725|M.bovis_AF2122/97 HRAAAPGASYYVLVFAKGAFPAELEVKP--NEVDEDELRAAVSKYWKIDE
Rv3699|M.tuberculosis_H37Rv HRAAAPGASYYVLVFAKGAFPAELEVKP--NEVDEDELRAAVSKYWKIDE
MMAR_5210|M.marinum_M HRAAAPGASYFVLVFAKGAFPAALETKP--NEVDEDELRAAVGKYWEIDE
MAV_0404|M.avium_104 HRAAAAGASLFILVFAKGAFPAEMQPKP--NEVDEDELRAAVGKYWVIDE
MSMEG_6235|M.smegmatis_MC2_155 HRASAPGARYFVLVFAKGAFPADLEVKP--NEVTEEELRTTVGKYWVVDD
TH_1722|M.thermoresistible__bu HRAAAPGARYFALVFAKGAFPEEMEQGPGPNPVDEDELREAVSKYWEIDE
MAB_0961c|M.abscessus_ATCC_199 SRAAAPDASYFVLVFAVGAFPPGI--GP--NAVTEDELRAAVEPYWAIDE
MUL_0662|M.ulcerans_Agy99 VRAAAPGASYFVLVFDKAALSGDG---P-INGVTEEELRDAVSKHWVIDE
**: ..* : *** .*:. * * * * *** :* :* :*:
Mflv_1324|M.gilvum_PYR-GCK IRPALIHTN----VPKLPGMP-PPPLDML-----DDRGHLR------MQA
Mvan_5471|M.vanbaalenii_PYR-1 IRPALIDTN----VPKLPGMP-PPPLDML-----DDRGRLR------MQA
Mb3725|M.bovis_AF2122/97 IRPAFIHVNPVTIPPQLAGAPVEFPPYDH-----DEKGRVK------FPA
Rv3699|M.tuberculosis_H37Rv IRPAFIHVNPVTIPPQLAGAPVEFPPYDH-----DEKGRVK------FPA
MMAR_5210|M.marinum_M IRPAFIHANVPVIPPLPAGSPIEFPPHQR-----DEKGRMK------FPA
MAV_0404|M.avium_104 IRPSFIVSN----VPQIADAPFEFPAHER-----DERGRMK------MPA
MSMEG_6235|M.smegmatis_MC2_155 IRPAFIHANAV----VFPDAP-ELPPHGY-----DDKGRIM------FPA
TH_1722|M.thermoresistible__bu IRPAFIHAN------MPPDVP-GVESFDT-----DEKGRTK------LPA
MAB_0961c|M.abscessus_ATCC_199 LSSAYIHSN---IPEVIPGANFEMPVFHK-----DEKGRNK------QPA
MUL_0662|M.ulcerans_Agy99 IRPARIHAN---LPPEVPGCPPSTLATNPRAASPSVRGCSQPIWADGTPH
: .: * * .. . :*
Mflv_1324|M.gilvum_PYR-GCK FLLSAHKEP
Mvan_5471|M.vanbaalenii_PYR-1 FLLQAHKES
Mb3725|M.bovis_AF2122/97 YLLTAHKAG
Rv3699|M.tuberculosis_H37Rv YLLTAHKAG
MMAR_5210|M.marinum_M YLLTAHKA-
MAV_0404|M.avium_104 YLLTAHKAQ
MSMEG_6235|M.smegmatis_MC2_155 FLLEAHRD-
TH_1722|M.thermoresistible__bu FLLSAHKA-
MAB_0961c|M.abscessus_ATCC_199 FLLQAHKR-
MUL_0662|M.ulcerans_Agy99 AVYSPANEG
: . .