For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTAPDPTDPTRFEEMYRDQRVAHGLPAATPWDIGGPQPVVEQLVALGAVKGEVLDPGTGPGHHAIYYASK GYSATGIDGSATAIERARDNAAKAGVSVNFQVADATRLEGLDNRFDTVVDCAFYHTFGTAPELQRSYVRA LWRATKPGARLYMYEFGAHQVNGLTAIRSLPEENFREMLPLGGWEITYLGPTTYRTTVSMETLETMAARN PEVADQVGPLVERFKTIQPWLTDGIAHVPFWEVHATRLD
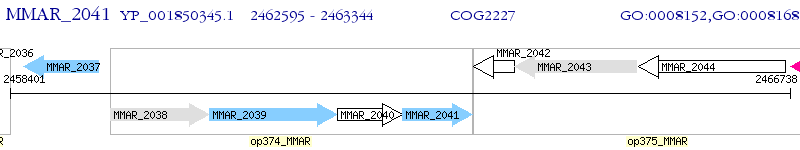
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2041 | - | - | 100% (249) | methyltransferase |
| M. marinum M | MMAR_2193 | - | 1e-29 | 42.16% (185) | NodS-like (sam)-dependent methyltransferase |
| M. marinum M | MMAR_0909 | - | 3e-22 | 31.94% (216) | methyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2694c | - | 1e-115 | 77.60% (250) | hypothetical protein Mb2694c |
| M. gilvum PYR-GCK | Mflv_4471 | - | 1e-26 | 34.30% (207) | methyltransferase type 11 |
| M. tuberculosis H37Rv | Rv2675c | - | 1e-115 | 77.60% (250) | hypothetical protein Rv2675c |
| M. leprae Br4923 | MLBr_01723 | - | 4e-05 | 32.32% (99) | hypothetical protein MLBr_01723 |
| M. abscessus ATCC 19977 | MAB_0240 | - | 2e-42 | 46.36% (220) | hypothetical protein MAB_0240 |
| M. avium 104 | MAV_3570 | - | 1e-110 | 73.88% (245) | thiopurine S-methyltransferase (tpmt) superfamily protein |
| M. smegmatis MC2 155 | MSMEG_3041 | - | 7e-30 | 40.91% (198) | thiopurine S-methyltransferase (tpmt) superfamily protein |
| M. thermoresistible (build 8) | TH_1722 | - | 8e-22 | 35.95% (153) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3309 | - | 1e-146 | 99.20% (249) | methyltransferase |
| M. vanbaalenii PYR-1 | Mvan_1888 | - | 3e-28 | 39.15% (189) | methyltransferase type 11 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2041|M.marinum_M --------------MTAP-DPTDPTRFEEMYRDQRVAHGLP--AATPWDI
MUL_3309|M.ulcerans_Agy99 --------------MTAP-DPTDPTRFEEMYRDQRVAHGLP--AATPWDI
Mb2694c|M.bovis_AF2122/97 --------------MTAQFDPADPTRFEEMYRDDRVAHGLP--AATPWDI
Rv2675c|M.tuberculosis_H37Rv --------------MTAQFDPADPTRFEEMYRDDRVAHGLP--AATPWDI
MAV_3570|M.avium_104 --------------MTASFDPADPARFEEMYRDQRTSHGLP--AATPWDI
MAB_0240|M.abscessus_ATCC_1997 ------------------MRPDGPGFFAELMYAVN---------AAPWDI
Mflv_4471|M.gilvum_PYR-GCK ---------------MDMDQTPTLSRFDDFYKDED--------KTPPWVI
Mvan_1888|M.vanbaalenii_PYR-1 -----------------MDLTPRLSRFDEFYKN----------QTPPWVI
MSMEG_3041|M.smegmatis_MC2_155 ----------------MTGPTGSEFDFDALYRGESPAEGVPPITSVPWDT
TH_1722|M.thermoresistible__bu ----------MATEDPGTGDPTDTIDWDSVYRQEAPGFDGP----PPWNI
MLBr_01723|M.leprae_Br4923 MTRSTEISANAVPNPHATAEQVAAARKDSKLAQVLYHDWEAENYDEKWSI
*
MMAR_2041|M.marinum_M GGPQ-----------PVVEQLVALGAVKGEVLDPGTGPGHHAIYYASKGY
MUL_3309|M.ulcerans_Agy99 GGPQ-----------PVVEQLVALGAVKGEVLDPGTGPGHHAIYYASKGY
Mb2694c|M.bovis_AF2122/97 GGPQ-----------PVVQQLVALGAIRGEVLDPGTGPGHHAIYYAAKGY
Rv2675c|M.tuberculosis_H37Rv GGPQ-----------PVVQQLVALGAIRGEVLDPGTGPGHHAIYYAAKGY
MAV_3570|M.avium_104 GGPQ-----------PVVRQLVALGAVKGEVLDPGTGPGHHAIYYASQGF
MAB_0240|M.abscessus_ATCC_1997 GGPQ-----------PVIRQLVALGAVRGEVLDPGCGTGWHAIEYARAGC
Mflv_4471|M.gilvum_PYR-GCK GEPQ-----------PAVVDIERAGLITGRVLDVGCGTGERTILLTAAGY
Mvan_1888|M.vanbaalenii_PYR-1 GEPQ-----------QAIVELEQAGLIGGRVLDVGCGTGEHTILLARAGY
MSMEG_3041|M.smegmatis_MC2_155 KAPK-----------ENVVAWEQAGLVHGHVLDIGCGLGDNAIYLAQQGY
TH_1722|M.thermoresistible__bu GEPQ-----------PELAALHRAGAFRSDVLDAGCGHAEISMALAADGY
MLBr_01723|M.leprae_Br4923 SYDQRCVDYVRHCFDAIVPDELFTELPYDCALELGCGTGFFLLNLIQSGV
: : . .*: * * . : *
MMAR_2041|M.marinum_M SATG--IDGSATAIERARDNAAKAGVSVN-FQVADATRLEGLDNRFDTVV
MUL_3309|M.ulcerans_Agy99 SATG--IDGSATAIERARDNAAKAGVSVN-FQVADATRLEGLDNRFDTVV
Mb2694c|M.bovis_AF2122/97 AATG--IDGSVAAIERARDNARKAGVSVN-FQVGDATTLDGLDGRFDTVV
Rv2675c|M.tuberculosis_H37Rv AATG--IDGSVAAIERARDNARKAGVSVN-FQVGDATTLDGLDGRFDTVV
MAV_3570|M.avium_104 SATG--IDGSAAAIERARANARTAGVSVN-FELADATKLDGFDGRFDTVV
MAB_0240|M.abscessus_ATCC_1997 SVTG--VDLAPTAIARARNNTRTAGVEAQ-FVLGDATTLD-YEARFDTVV
Mflv_4471|M.gilvum_PYR-GCK DVLG--VDGAPSAVEQARHNAKTRVVDAR-FDVADALDLG-DEPAYDTII
Mvan_1888|M.vanbaalenii_PYR-1 DVLG--IDGAPTAVEQARRNAEAQGVDAR-FELADALHLG-PDPTYDTIV
MSMEG_3041|M.smegmatis_MC2_155 RVTA--LDISPTALITAERRAADAGVDVT-FAVADATRLDGYDDVFDTVV
TH_1722|M.thermoresistible__bu TVVG--IDLSPTAIAAANRTAQERGLATASFVQGDITDFGGYDGRFNTII
MLBr_01723|M.leprae_Br4923 ARRGSVTDLSPGMVKIATRNGQSLGLDIN-GRVADAEGIPYDDNTFDLVV
. * : : * : .* : : :: ::
MMAR_2041|M.marinum_M DCAFYHTFGTAPELQRSYVRALWR------ATKPGAR--LYMYEFG----
MUL_3309|M.ulcerans_Agy99 DCAFYHTFGTAPELQRSYVQALWR------ATKPGAR--LYMYEFG----
Mb2694c|M.bovis_AF2122/97 DCAFYHTFSTAPELQRCYVRALRR------ASKPGAR--LYMFEFG----
Rv2675c|M.tuberculosis_H37Rv DCAFYHTFSTAPELQRCYVRALRR------ASKPGAR--LYMFEFG----
MAV_3570|M.avium_104 DCAFYHVFATEPELRSSYARALHR------ATKPGAR--LYMFEFG----
MAB_0240|M.abscessus_ATCC_1997 DSKLLDNLDSA-EARGRYLRSLYR------AMKPQGR--LFLYGFS----
Mflv_4471|M.gilvum_PYR-GCK DSALFHIFDDA--DREKYVRSLGR------ATAPGSV--VHLLALSD---
Mvan_1888|M.vanbaalenii_PYR-1 DSALFHIFDDA--DRATYVRSLHA------ATRPGSV--VHLLALSD---
MSMEG_3041|M.smegmatis_MC2_155 DSGMFHCLSDD--AKGSYAAAVHR------ATRAGAT--LLLGAFSD---
TH_1722|M.thermoresistible__bu DSTLFHSIPVQ--ARDGYLRSIHR------AAAPGAR--YFALVFAKG--
MLBr_01723|M.leprae_Br4923 GHAVLHHIPDVELSLREVLRVLKPGGRFVFAGEPTDVGNRYARVFADLTW
. . . : : * . :.
MMAR_2041|M.marinum_M ---------AHQVNGLTAIRSLPEEN----FREMLPLGGWEITYLGPTTY
MUL_3309|M.ulcerans_Agy99 ---------AHQVNGLTAIRSLPEEN----FREMLPLGGWEITYLGPTTY
Mb2694c|M.bovis_AF2122/97 ---------EHNVNGFSMPRSLSEDD----FRQVLPVGGWEITYLGTTTY
Rv2675c|M.tuberculosis_H37Rv ---------EHNVNGFSMPRSLSEDD----FRQVLPVGGWEITYLGTTTY
MAV_3570|M.avium_104 ---------EHDVNGFKMMRSLSEND----FRDVLPAAGWEISYLGPTTY
MAB_0240|M.abscessus_ATCC_1997 ---------DGHVNGFHN-HELDGID----YEMALTAAGFTITYLGETTY
Mflv_4471|M.gilvum_PYR-GCK ---------TGRGFG----PEVSEET----IRAAFADPGWDIEALTTTYY
Mvan_1888|M.vanbaalenii_PYR-1 ---------SGRGFG----PEVSEHT----IRAAFG-AGWEVEALTETTY
MSMEG_3041|M.smegmatis_MC2_155 ---------ANLQHDNWQAAWVSEDT----LRSSLDDGGWDITSLTTETI
TH_1722|M.thermoresistible__bu -------AFPEEMEQGPGPNPVDEDE----LREAVS-KYWEIDEIRPAFI
MLBr_01723|M.leprae_Br4923 KVTIRVVQLPGLSAWRRTQVEIDENSRAAALEWVVDLHTFEPKVLETMAT
: . . : :
MMAR_2041|M.marinum_M RTTVSMETLETMAARNPEVADQVGPLVERFKTIQP-----------WLTD
MUL_3309|M.ulcerans_Agy99 RTTVSMETLETMAARNPEVADQVGPLVERFKTIQP-----------WLTD
Mb2694c|M.bovis_AF2122/97 QVNLSVEALELMAARNPDMADQVRCVLERFRAIKP-----------WLVG
Rv2675c|M.tuberculosis_H37Rv QVNLSVEALELMAARNPDMADQVRCVLERFRAIKP-----------WLVG
MAV_3570|M.avium_104 QVNLSAETIQMMAARNPDMADEAAKLLERFRAMEP-----------WLQA
MAB_0240|M.abscessus_ATCC_1997 R--LSIPSYRRICTDCP---DQVP------------------------PD
Mflv_4471|M.gilvum_PYR-GCK RGVITAPHADAMGLDVG---------------------------------
Mvan_1888|M.vanbaalenii_PYR-1 RGVVIDAHTEALNLPAG---------------------------------
MSMEG_3041|M.smegmatis_MC2_155 R-VPGAALAGSGDEGGA---------------------------------
TH_1722|M.thermoresistible__bu HANMPPDVPGVESFDTDE-------------------------------K
MLBr_01723|M.leprae_Br4923 NAGAVQVKTVTEEFTAAMLGWPLRTFESTMPPSKLGWGWARFAFTSWKTL
.
MMAR_2041|M.marinum_M GIAHVPFWEVHATRLD------------
MUL_3309|M.ulcerans_Agy99 GIAHVPFWEVHATRPD------------
Mb2694c|M.bovis_AF2122/97 GRVHAPFWEVHATRVD------------
Rv2675c|M.tuberculosis_H37Rv GRVHAPFWEVHATRVD------------
MAV_3570|M.avium_104 GRVHAPFWEVHATRVD------------
MAB_0240|M.abscessus_ATCC_1997 GQIHIPMIEIHAVRNGLWQA--------
Mflv_4471|M.gilvum_PYR-GCK TVVDEPAWLARIRRL-------------
Mvan_1888|M.vanbaalenii_PYR-1 TVVDEPAWSARIRRL-------------
MSMEG_3041|M.smegmatis_MC2_155 DEFDAAFFLVQAQRR-------------
TH_1722|M.thermoresistible__bu GRTKLPAFLLSAHKA-------------
MLBr_01723|M.leprae_Br4923 SWVDANVWRRVIPKSWFYNIIITGVKPS
. :