For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PRKSATFRKVPTQDRSKDTVKRILDAARMLLEERGYDGTSTNRIAAAAGISPGSLYQYFANKDAILHAVV AEYTQQLLDQVTGNLASLVRSEHDVPLSAAIEAQVDAMLERPQILRVISGQLPGRTSADVLRPLRELMTA TIKGYAIAAPDQSIDMDVDAATWIIVQLLGSTIRYVVDEPPIAKDVFVTEMTRFVMSSPVAWRCSARPAA VPTA*
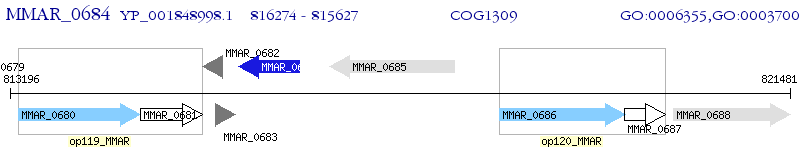
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0684 | - | - | 100% (215) | putative regulatory protein |
| M. marinum M | MMAR_4832 | - | 7e-11 | 33.33% (117) | TetR family transcriptional regulator |
| M. marinum M | MMAR_3161 | - | 3e-10 | 26.77% (198) | transcriptional regulatory protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0149 | - | 2e-10 | 28.90% (173) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_3251 | - | 3e-16 | 27.03% (185) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0144 | - | 3e-10 | 28.90% (173) | TetR family transcriptional regulator |
| M. leprae Br4923 | MLBr_02677 | - | 5e-06 | 33.03% (109) | putative TetR-family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_3372 | - | 1e-15 | 28.35% (194) | TetR family transcriptional regulator |
| M. avium 104 | MAV_0110 | - | 5e-10 | 30.00% (140) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_6532 | - | 4e-14 | 27.23% (191) | transcriptional regulatory protein |
| M. thermoresistible (build 8) | TH_3831 | - | 6e-11 | 30.06% (163) | PUTATIVE putative transcriptional regulator, TetR family |
| M. ulcerans Agy99 | MUL_4386 | - | 5e-09 | 30.07% (153) | hypothetical protein MUL_4386 |
| M. vanbaalenii PYR-1 | Mvan_0955 | - | 2e-30 | 35.32% (201) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0149|M.bovis_AF2122/97 --------------------------------------------------
Rv0144|M.tuberculosis_H37Rv --------------------------------------------------
TH_3831|M.thermoresistible__bu --------------------------------------------------
MAB_3372|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6532|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0955|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0684|M.marinum_M --------------------------------------------------
Mflv_3251|M.gilvum_PYR-GCK --------------------------------------------------
MUL_4386|M.ulcerans_Agy99 --------------------------------------------------
MAV_0110|M.avium_104 MKRRPKDRKVQIARAATDAFSELGYHAVSMENIAARVGISAAALYRHSQG
MLBr_02677|M.leprae_Br4923 --------------------------------------------------
Mb0149|M.bovis_AF2122/97 --------------------------------------------------
Rv0144|M.tuberculosis_H37Rv --------------------------------------------------
TH_3831|M.thermoresistible__bu --------------------------------------------------
MAB_3372|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6532|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0955|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0684|M.marinum_M --------------------------------------------------
Mflv_3251|M.gilvum_PYR-GCK --------------------------------------------------
MUL_4386|M.ulcerans_Agy99 --------------------------------------------------
MAV_0110|M.avium_104 KYDLFREVFLALGQQLVDATAFADDLPADADPGETLSALTTALIDTTIVN
MLBr_02677|M.leprae_Br4923 --------------------------------------------------
Mb0149|M.bovis_AF2122/97 ------------------------MPHSWTPTSVMTPPLVVAAFRPVGHY
Rv0144|M.tuberculosis_H37Rv ------------------------MPHSWTPTSVMTPPLVVAAFRPVGHY
TH_3831|M.thermoresistible__bu --------------------------------------------------
MAB_3372|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6532|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0955|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0684|M.marinum_M --------------------------------------------------
Mflv_3251|M.gilvum_PYR-GCK --------------------------------------------------
MUL_4386|M.ulcerans_Agy99 --------------------------------------------------
MAV_0110|M.avium_104 RTAGGLYRWEGRYLRGDDQRKLAEQIKVINRRLQRPLIKLRPKLTSRQRW
MLBr_02677|M.leprae_Br4923 --------------------------------------------------
Mb0149|M.bovis_AF2122/97 RLATDRAGGPCSPPATGAKLTSSVASRPTVGTKPQWWHTLVMSMSLTAGR
Rv0144|M.tuberculosis_H37Rv RLATDRAGGPCSPPATGAKLTSSVASRPTVGTKPQWWHTLVMSMSLTAGR
TH_3831|M.thermoresistible__bu --------------------------------------------------
MAB_3372|M.abscessus_ATCC_1997 -------------------------------MAILAFGEGFDIMSSSRLQ
MSMEG_6532|M.smegmatis_MC2_155 ------------------------------------------MVVAMSDA
Mvan_0955|M.vanbaalenii_PYR-1 ------------------------------------------MTSDVASR
MMAR_0684|M.marinum_M -------------------------------------------MPRKSAT
Mflv_3251|M.gilvum_PYR-GCK -----------------------------------------MSAETRSSG
MUL_4386|M.ulcerans_Agy99 ----------------------------------------MSDRVTAAVE
MAV_0110|M.avium_104 ILSAATLSVIGSITDHRSRLGNAEIRATLSDIAAAVLKAELPTQRGRGSE
MLBr_02677|M.leprae_Br4923 ------------------------------------------------ME
Mb0149|M.bovis_AF2122/97 GPGRPPAAKADETRKRILHAARQVFSERGYDGATFQEIAVRADLTRPAIN
Rv0144|M.tuberculosis_H37Rv GPGRPPAAKADETRKRILHAARQVFSERGYDGATFQEIAVRADLTRPAIN
TH_3831|M.thermoresistible__bu --MRPARTASSDTRARILEAARRIIVERGYPAATCQGIARAAGVSRPTLH
MAB_3372|M.abscessus_ATCC_1997 PRKQPRQDRAEATRRRILEAAAHVFADFGYAAGTTNRIAERAEISIGSLY
MSMEG_6532|M.smegmatis_MC2_155 PRKKPRQRRSRETVEVVLEAAAQVFDREGLAA-TTNRIAERAGVSIGSLY
Mvan_0955|M.vanbaalenii_PYR-1 LRRAPRQERSRAMVDRILDAGEQMLIAHGFDGASTNRIAAAAGISPGSLY
MMAR_0684|M.marinum_M FRKVPTQDRSKDTVKRILDAARMLLEERGYDGTSTNRIAAAAGISPGSLY
Mflv_3251|M.gilvum_PYR-GCK RRKLPKQERSREMVAKIVQTTAALLIKTEPEQVTTNLIADRAGISKGSIY
MUL_4386|M.ulcerans_Agy99 RTLDDRQREATEEVERILAAAVRVMERAAPEPPRVSDIVAEAGSSNKAFY
MAV_0110|M.avium_104 PARAPTLTSAAGSYELLLHESMRLFNERGYRETGMEDIAAAVGIPVASIY
MLBr_02677|M.leprae_Br4923 SQRRTQEERSTATRDALTLARHRLWRLRGYAEVGTPEIAKEADTTRGAVY
: : : *. . . :.
Mb0149|M.bovis_AF2122/97 HYFANKRVLYQEVVEQTHELVIVAGIERARRE--PTLMGRLAVVVDFAME
Rv0144|M.tuberculosis_H37Rv HYFANKRVLYQEVVEQTHELVIVAGIERARRE--PTLMGRLAVVVDFAME
TH_3831|M.thermoresistible__bu YYFASREEIYDCLVEQSRNLLAEC-AERAMRY--DTLVERLTEFGLRLRE
MAB_3372|M.abscessus_ATCC_1997 QYFPNKDAILVELMTAHTHRGFALIHQHLAAG--LPDT--IEDTVRVFVG
MSMEG_6532|M.smegmatis_MC2_155 QYFPNKHALLNALAQRHVQTASARLDAVFAEL--RHETPEFDQTLRSILA
Mvan_0955|M.vanbaalenii_PYR-1 QYFPNKDAIAAAVIDRFSDQLSARVAARVSER--LDQP--APDYVRESIA
MMAR_0684|M.marinum_M QYFANKDAILHAVVAEYTQQLLDQVTGNLASL--VRSEH--DVPLSAAIE
Mflv_3251|M.gilvum_PYR-GCK QYFSDKEEIIEAAVEHLAAQQAPEIEEMLRSV--TMSRP--EVAMESSID
MUL_4386|M.ulcerans_Agy99 RYFAGKDDLILAVMERGVAIVVSYLGHQMAKE--SRPDTKIARWIEGTLA
MAV_0110|M.avium_104 QYFPGKAAILAVYYRRAADQLSADLSSILATS--SDPE----QALARLIE
MLBr_02677|M.leprae_Br4923 HQFTDKAALFRDVVETVEQDAMAQIASVVASSGAKTPVDAIRAKVDAWLE
*..: :
Mb0149|M.bovis_AF2122/97 ADAQYPASTAFLATTVLES------QRHPELSRTENDAVRATREFLVWAV
Rv0144|M.tuberculosis_H37Rv ADAQYPASTAFLATTVLES------QRHPELSRTENDAVRATREFLVWAV
TH_3831|M.thermoresistible__bu VDSRDRSTIAFLISARLEG------MRNRELG---EDPAMALREFFTELL
MAB_3372|M.abscessus_ATCC_1997 SAIANHRDSPRLHRVLFEE------APRPPEF--LAMLHRSEDAVIAAAE
MSMEG_6532|M.smegmatis_MC2_155 VVVDLHHDRPALHRLMHRV------AVPRDDQ--LAALQEFENRLAGEVA
Mvan_0955|M.vanbaalenii_PYR-1 ALIEALDVHPEFLRAVMEQ------TPRLGTG--SKLVAFEQRIGELTVA
MMAR_0684|M.marinum_M AQVDAMLERPQILRVISGQ------LPGRTSA---DVLRPLRELMTATIK
Mflv_3251|M.gilvum_PYR-GCK ILIDFTIANRRLIRYLAER------PDHVRTF--EAISGLNATLLAMATL
MUL_4386|M.ulcerans_Agy99 QVADPHLIS--MSRAAAGQ------LSN---------WLATQREMLRPLR
MAV_0110|M.avium_104 AYVTRSFANPELACVYYTE------RHNLPET-DAVLLYNIQRSTVESWA
MLBr_02677|M.leprae_Br4923 GLADPQVRQLILLYAPSVLGWAAFRDVAQHYSLGMTEQQLLSEASRARQL
:
Mb0149|M.bovis_AF2122/97 NDAIERGELAADVDVSSLAETLLVVLCGVGFYIGFVGSYQRMATITDSFQ
Rv0144|M.tuberculosis_H37Rv NDAIERGELAADVDVSSLAETLLVVLCGVGFYIGFVGSYQRMATITDSFQ
TH_3831|M.thermoresistible__bu ADAVAAGELPSDIDIESAVEVMYTVLWGLGFYTGFVERSTDMLVLAEQLE
MAB_3372|M.abscessus_ATCC_1997 AVLAAHPEVR-VTETDTAARMVVATIESLVHRLIAGPRPIDPQLFEDQLV
MSMEG_6532|M.smegmatis_MC2_155 FHLVRCGRVRDPDDASATARTVVHAVDAHLHRVLP-HREVTREAAVDDLV
Mvan_0955|M.vanbaalenii_PYR-1 YLTINKRQVRTDVTVDVAAWMLVRMVEHLCVRYILDQPSIEREKFIDEVT
MMAR_0684|M.marinum_M GYAIAAPDQSIDMDVDAATWIIVQLLG-STIRYVVDEPPIAKDVFVTEMT
Mflv_3251|M.gilvum_PYR-GCK HMGHYRDQYRDELSPRALAWLFFNMAVATTLRYIEADDPISLDELRSGLK
MUL_4386|M.ulcerans_Agy99 FLLTAPVTALGSSDVDRDVEAVYGCTVATMRRYVGSAERPGRDDIEHLVR
MAV_0110|M.avium_104 RLAVAARPELTLGRARYAVHAAFALAVDIGRLVYPNTGAPASTTVRRLME
MLBr_02677|M.leprae_Br4923 AQPLDHVLIGKLYESAMAIRRHHRLPEASPARNQTGAAPTHREHVARPSC
Mb0149|M.bovis_AF2122/97 QLLAGTLWRPPT----------------------
Rv0144|M.tuberculosis_H37Rv QLLAGTLWRPPT----------------------
TH_3831|M.thermoresistible__bu NLFASA----------------------------
MAB_3372|M.abscessus_ATCC_1997 DMLTRYLTG-------------------------
MSMEG_6532|M.smegmatis_MC2_155 DLVERLLRP-------------------------
Mvan_0955|M.vanbaalenii_PYR-1 LMALNYLRPWPTPPEDCFGSPR------------
MMAR_0684|M.marinum_M RFVMSSPVAWRCSARPAAVPTA------------
Mflv_3251|M.gilvum_PYR-GCK FASTGLLAGAVSGGHVGEPGPS------------
MUL_4386|M.ulcerans_Agy99 FCLLGLGVS-------------------------
MAV_0110|M.avium_104 VTVLGRPAAASRDGLRGAGRQRR-----------
MLBr_02677|M.leprae_Br4923 EALTRHIRHRSTRTRSHRGKRGELLPRAATSSLL