For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MADTIVGVLDEAAASRPARPLLRDCDGATLTVAETAALSAAGTRWLWDAGIRPGMTIAWQLPSHVSAALL MLALARCSVTQAPVLHLYRQREACAALAVAGVDTLIVDESTTANVPPGARVVKLPDGFVELLRALPTDPQ PELAQSHRADDARWIYFTSGTTGRPKGVRHTDATLLAAARGYAQYLGLGEHPNEVGTIAFPIAHIGGMVY LASALIADFPVLMVPKVDVETLPRLLAQHDVTVTGASTAYYQMLLSAQVASGSGEPVVPSLRMLIGGGAA CPPELHKQVREQMGVPIVHAYGMTEAAMVCVSPATDTEEQQSNSSGRPIPGSDVRISETGEVELRGANLT PGYVDAEQWKSVLTSDGWFRTGDRGYLRPDGRIVITGRTKDLIIRKGENIAPDEIENELLAHPLVDEVAV VGQPDDLRGEMVCAVVRRSPRHRDVTLDELCAFLDQRGMMKQKWPERLFFVDEFPLTGLGKVAKAELVQQ ISGGRR*
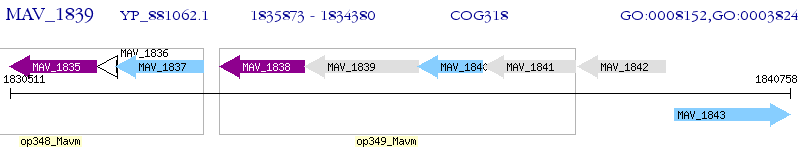
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1839 | - | - | 100% (497) | cyclohex-1-ene-1-carboxylate:CoA ligase |
| M. avium 104 | MAV_2571 | - | 0.0 | 71.34% (499) | cyclohex-1-ene-1-carboxylate:CoA ligase |
| M. avium 104 | MAV_2116 | - | 1e-80 | 36.33% (523) | AMP-binding enzyme, putative |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2405 | mbtA | 9e-40 | 29.14% (532) | bifunctional salicyl-AMP ligase/salicyl-S-arcp synthetase |
| M. gilvum PYR-GCK | Mflv_1784 | - | 1e-56 | 32.87% (505) | AMP-dependent synthetase and ligase |
| M. tuberculosis H37Rv | Rv2384 | mbtA | 9e-40 | 29.14% (532) | bifunctional salicyl-AMP ligase/salicyl-S-arcp synthetase |
| M. leprae Br4923 | MLBr_02661 | - | 2e-32 | 26.60% (530) | acyl-CoA synthetase |
| M. abscessus ATCC 19977 | MAB_2247c | - | 3e-42 | 28.38% (525) | bifunctional salicyl-AMP ligase/salicyl-S-arcp synthetase |
| M. marinum M | MMAR_0303 | - | 8e-50 | 30.02% (503) | acyl-CoA synthetase |
| M. smegmatis MC2 155 | MSMEG_4774 | - | 1e-47 | 31.34% (485) | AMP-dependent synthetase and ligase |
| M. thermoresistible (build 8) | TH_3528 | - | 0.0 | 70.56% (496) | PUTATIVE cyclohex-1-ene-1-carboxylate:CoA ligase |
| M. ulcerans Agy99 | MUL_3145 | - | 2e-38 | 29.50% (478) | hypothetical protein MUL_3145 |
| M. vanbaalenii PYR-1 | Mvan_4966 | - | 7e-57 | 33.80% (500) | AMP-dependent synthetase and ligase |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_1839|M.avium_104 -------------------------------------MMADTIVGVLDEA
TH_3528|M.thermoresistible__bu --------------------------------------MTATIAGLLDDA
Mflv_1784|M.gilvum_PYR-GCK --------------------------------------MTGTLVEALRDA
Mvan_4966|M.vanbaalenii_PYR-1 --------------------------------------MT-TLADALRDA
MMAR_0303|M.marinum_M -----MGGPKVPQRRLIDGRWVRWDADRAAGAYAHRWWVRRTLADALREA
MSMEG_4774|M.smegmatis_MC2_155 --------------------MRDIPAELTKRYVEEGWWTPETLGQMLAEG
Mb2405|M.bovis_AF2122/97 -MPPKAADGRRPSPDGGLGGFVPFPADRAASYRAAGYWSGRTLDTVLSDA
Rv2384|M.tuberculosis_H37Rv -MPPKAADGRRPSPDGGLGGFVPFPADRAASYRAAGYWSGRTLDTVLSDA
MAB_2247c|M.abscessus_ATCC_199 MTQPRSTTTASSAATPALDGFVPFPSVRADEYRRAGYWTGNTVESLLRVS
MUL_3145|M.ulcerans_Agy99 --------------------------------------------------
MLBr_02661|M.leprae_Br4923 --------------MKSESITSSRAPVEQIDARLTGQFESPSIASLVEAA
MAV_1839|M.avium_104 AASRPARPLLRDCD--GATLT--VAETAALSAAGTRWLWDAGIRPGMTIA
TH_3528|M.thermoresistible__bu AARKPDQRMLIDVD--GGCLT--VAEVARLSSAATRWLADAGVQPGMTVA
Mflv_1784|M.gilvum_PYR-GCK ARDTPDRVLVRDDA--VALTC---AELLSAATGLAHALTRR-MPAGSVVS
Mvan_4966|M.vanbaalenii_PYR-1 ARDTPDRILLRDDE--ASLSS---AELLSAATNLAHALVRR-MPVGSVVS
MMAR_0303|M.marinum_M AQSTPQRVALVDAD--RQLDC---QELHRQAMALAQHLRSR-MPAGSVVS
MSMEG_4774|M.smegmatis_MC2_155 LAAGPDVVFCVHSE--VRPYTGTFGEVETLARRLAAGLRARGVGPGDVIA
Mb2405|M.bovis_AF2122/97 ARRWPDRLAVADAGDRPGHGGLSYAELDQRADRAAAALHGLGITPGDRVL
Rv2384|M.tuberculosis_H37Rv ARRWPDRLAVADAGDRPGHGGLSYAELDQRADRAAAALHGLGITPGDRVL
MAB_2247c|M.abscessus_ATCC_199 AARRPCHPAVIDE-----HRALSYRELDEAADNAAHAFLRLGIAPGDRVL
MUL_3145|M.ulcerans_Agy99 ------------------------------------------MLPGDRVA
MLBr_02661|M.leprae_Br4923 AIRNPSAPALVVTDN---RIVVSYRDLLRLVDDLTVQLALGGLLPGDRVA
: * :
MAV_1839|M.avium_104 WQLPSHVSAALLMLALARCSVTQAPVLHLYRQREACAALAVAGVDTLIVD
TH_3528|M.thermoresistible__bu WQLPAHSAAALLMLALARMPVVQAPVLHLYRQREVQAALDVAGADVLIVD
Mflv_1784|M.gilvum_PYR-GCK FTVPNWHEAAIIYLGATLAGMVVNPILPSLRERELSFILDDADSRMIFVP
Mvan_4966|M.vanbaalenii_PYR-1 FAVPNWHEAAVIYLGATLAGMVVNPILPSLRERELSFILADAGSRAIFIP
MMAR_0303|M.marinum_M FMLPNWHEAAVIYLASTLAAMVANPVLPSLREHELRFILEDAGSRMIFAP
MSMEG_4774|M.smegmatis_MC2_155 FQLPNWMEAAAAFWATSFLGAVTVPIVHFYGRKELAHILSTAKPKVFLTT
Mb2405|M.bovis_AF2122/97 LQLPNGCQFAVALFALLRAGAIPVMCLPGHRAAELGHFAAVSAATGLVVA
Rv2384|M.tuberculosis_H37Rv LQLPNGCQFAVALFALLRAGAIPVMCLPGHRAAELGHFAAVSAATGLVVA
MAB_2247c|M.abscessus_ATCC_199 LQLPNRASFAVALFGLMRAGAIPVMCLPGHRAAELSHFIEVADAVGLVIP
MUL_3145|M.ulcerans_Agy99 VWHGDCAALHALFIAIERCGAVVVGIGARAGVREATQILRAAQVKLLVSD
MLBr_02661|M.leprae_Br4923 LCAASNIEFVVGLLAASRAGLIVVPLDPALPVNEQCIRSQAAGVRVTLVD
. * : .
MAV_1839|M.avium_104 ESTTANVPPGARVVKLP----------------DGFVELLRALPTDPQPE
TH_3528|M.thermoresistible__bu ASTAANAPDDTSVITLP----------------DDFVDVLRAMPAEP--V
Mflv_1784|M.gilvum_PYR-GCK ESFGGHDYAAMLDRVAPGLPSPPEVVVVRGE---GRVTLADLLA-EPADG
Mvan_4966|M.vanbaalenii_PYR-1 QTFGGHDYAAMLDRVVAGLPSPPEVVVVRGEP--RRPALTDLVS-EPRDG
MMAR_0303|M.marinum_M ADFRGHDYTSMLRRVAAQLPSRPEVIEVRGQCPDGVSTFQSLLVGRETAA
MSMEG_4774|M.smegmatis_MC2_155 ERFGRMSFQPDLCAQVP-------IVGLVGES-----SFTDLLAERPMSG
Mb2405|M.bovis_AF2122/97 DVASGFDYRPMARELVADHPTLRHVIVDGDPG--PFVSWAQLCAQAGTGS
Rv2384|M.tuberculosis_H37Rv DVASGFDYRPMARELVADHPTLRHVIVDGDPG--PFVSWAQLCAQAGTGS
MAB_2247c|M.abscessus_ATCC_199 DSAGGFDYRALAVALSQRHPRLRTVVVEGDAG--PFASWTSLLKSVEPQG
MUL_3145|M.ulcerans_Agy99 APRSAAATEVSAQLPVPSGVLVREGKTLRFDG--KPVPVAPENGLESELG
MLBr_02661|M.leprae_Br4923 SLALEGVSDQRAATMRYWPIAVSYGSVTGASEGSLLVHLDDTAALHPVTS
MAV_1839|M.avium_104 LAQSHRADDARWIYFTSGTTGRPKGVRHTDATLLAAARGYAQYLGLGEHP
TH_3528|M.thermoresistible__bu LVEDPRPDDVRWIYFTSGTTGRPKGVRHTDSTLLAAARGFTAHLGLGTHP
Mflv_1784|M.gilvum_PYR-GCK VLPAPRPTDTRMILYTSGTTGRPKGVLHTHESLAALIAQLGRFWRI--DR
Mvan_4966|M.vanbaalenii_PYR-1 TLPDLDPAATRMILYTSGTTGRPKGVLHSHKSLGALIAQLGRYWRI--DP
MMAR_0303|M.marinum_M PLPTADPDAVRMVLYTSGTTGRPKGVLHTHNSIHALICQIRDHWMV--EP
MSMEG_4774|M.smegmatis_MC2_155 TLDT-DPAAPALIAFTSGTTRDPKGVIHSHQTLSCETRQLLANYPP--NR
Mb2405|M.bovis_AF2122/97 PAPPADPGSPALLLVSGGTTGMPKLIPRTHDDYVFNATASAALCRLS--A
Rv2384|M.tuberculosis_H37Rv PAPPADPGSPALLLVSGGTTGMPKLIPRTHDDYVFNATASAALCRLS--A
MAB_2247c|M.abscessus_ATCC_199 PLPTVQTDTPALLLVSGGTTAAPKLIPRTHEDYVYNATQAARVCGLT--Q
MUL_3145|M.ulcerans_Agy99 NERRLGPDDVFLINSTSGTTGAPKCVVHTQNRWHYFHQHAVANGMLT--P
MLBr_02661|M.leprae_Br4923 TPDGLRHDD-AMIMFTGGTTGLPKMVPWTDGNIAGSVHAIITAYQLG--P
: :.*** ** : :.
MAV_1839|M.avium_104 NEVGTIAFPIAHIGGMVY--LASALIADFPVLMVP--KVDVETLPRLLAQ
TH_3528|M.thermoresistible__bu QEVSTIAFPVAHIGGIVY--LSCALLAGFPVLLIP--KVEADELLRLLAE
Mflv_1784|M.gilvum_PYR-GCK GDTFLVPSPIAHIGGSIYA-FECPLLLGTEAVLMQ--RWDPDAAVGLMLE
Mvan_4966|M.vanbaalenii_PYR-1 GDTFLVPSPIAHIGGSIYA-FECPLLLGTQAVLMQ--RWDPDAAVALMLR
MMAR_0303|M.marinum_M GDGFLVASPIAHIGGSIYA-FECPLLLGTTAVLLD--RWDADRAVQLMTS
MSMEG_4774|M.smegmatis_MC2_155 G-RQLTATPVGHFIGMVGA-FLIPVLEGAPVDLCD--VWDPAKVLRLMES
Mb2405|M.bovis_AF2122/97 DDVYLVVLAAGHNFPLACPGLLGAMTVGATAVFAP--DPSPEAAFAAIER
Rv2384|M.tuberculosis_H37Rv DDVYLVVLAAGHNFPLACPGLLGAMTVGATAVFAP--DPSPEAAFAAIER
MAB_2247c|M.abscessus_ATCC_199 DDVYLVALPAGHNFPLACPGLLGAISTGATTVFLS--DPSPESAFATIAR
MUL_3145|M.ulcerans_Agy99 QDVVLPVIPMPFGFGIWTS-HTTPIYLGASAVILD--RFTTLAACEAIAR
MLBr_02661|M.leprae_Br4923 QDATVVVMPLYHGHGLIAA-LLSTLASGGVVLLPARARFSARTFWDDIDA
. . .: . . : :
MAV_1839|M.avium_104 HDVTVTGASTAYYQMLLSAQVASGSGEPVVPSLRMLIGGGAACPPELHKQ
TH_3528|M.thermoresistible__bu YRVTVTGGSTAFYQMLLSAQLASGQNEPLVPTLRMLIGGGAPCPPEVHRK
Mflv_1784|M.gilvum_PYR-GCK HRVTHMAGATPFLVGLLAAAQRAGTR---LPDLKVFICGGASVPPTLIRS
Mvan_4966|M.vanbaalenii_PYR-1 HRCTHMAGATPFLDGLLAAAERAGTR---LPDLKVFICGGASVPPSLIRR
MMAR_0303|M.marinum_M KRCTHMAGATPFLEQLLAAAQRAGTR---LPDLKFFVCGGASVSPSLIRC
MSMEG_4774|M.smegmatis_MC2_155 DGLSIGGGPPYFVTSLLDHPDFADRH---LARIRTVGLGGSTVPAAVTRR
Mb2405|M.bovis_AF2122/97 HGVTVTALVPALAKLWAQSCEWEPVTP---KSLRLLQVGGSKLEPEDARR
Rv2384|M.tuberculosis_H37Rv HGVTVTALVPALAKLWAQSCEWEPVTP---KSLRLLQVGGSKLEPEDARR
MAB_2247c|M.abscessus_ATCC_199 HRVSVTAVVPSLAQLWAQATAWEPVLP---ETLRLLQVGGAKLGADDARA
MUL_3145|M.ulcerans_Agy99 HRVTVLCCVSTQLTMLMADHWCRAYD---LSSLRVVFAGGEALPYRRATE
MLBr_02661|M.leprae_Br4923 VAATWYTAAPAIHRILLELASTQSFRSK-RAKLRFIRSCSAPLTQETAQA
: . :: . .
MAV_1839|M.avium_104 VREQMG-VPIVHAYGMTEAAMVCVSP-----ATDTEEQQSNSSGRPIPGS
TH_3528|M.thermoresistible__bu VRAHMG-IPIVHAYGMTEAPMIGVSE-----AADTEEQQSNSSGRPIPGS
Mflv_1784|M.gilvum_PYR-GCK AAAYFDKAAVSRVYGSTEVPVTTVG--------ALDDIDRAADTDGRPGI
Mvan_4966|M.vanbaalenii_PYR-1 AADYFDKAAVSRVYGSTEVPVTTVG--------SLDDVDRAADTDGRPGI
MMAR_0303|M.marinum_M ATDYFDKAVVTRVYGSTEVPVTTIG--------APDSSAHAAATDGRVGA
MSMEG_4774|M.smegmatis_MC2_155 LADLG--IFVFRSYGSTEHPSITGSS-----PSAPEGKRLYTDGNPRPGV
Mb2405|M.bovis_AF2122/97 VRTALT-PGLQQVFGMAEGLLNFTRI-----GDPPEVVEHTQGRPLCPAD
Rv2384|M.tuberculosis_H37Rv VRTALT-PGLQQVFGMAEGLLNFTRI-----GDPPEVVEHTQGRPLCPAD
MAB_2247c|M.abscessus_ATCC_199 VRESLT-QGLQQVFGMAEGLLCLTQP-----GDPTDILDNSQGRPMCDAD
MUL_3145|M.ulcerans_Agy99 FEDLTG-AAILQFYGSNETGLLSATT-----LRDSRERRLRTGGRVVPEM
MLBr_02661|M.leprae_Br4923 LREEFL-APVICAFGMTEATHQVTTTNIKWFGQGENPTVTNGLVGQSTGV
: :* *
MAV_1839|M.avium_104 DVRIS----------ETGEVELRGANLTPGYVDAEQWKS-VLTSDGWFRT
TH_3528|M.thermoresistible__bu EIRIA----------ANGEIXLRGSNVTTGYVX-DQWSG-STTADGWFRT
Mflv_1784|M.gilvum_PYR-GCK AEVRI----------VDGEIRARGPQMFTGYLHPEDDRE-SFDDAGYFRT
Mvan_4966|M.vanbaalenii_PYR-1 ADVTL----------VDGEIRARGPQMLTGYLHPEDNSE-SFDAEGFFRT
MMAR_0303|M.marinum_M ADVKL----------VAGEIRVRGPQMLVGYRHPEDEAG-SFDAQGYFRT
MSMEG_4774|M.smegmatis_MC2_155 EIRLG----------PDGEIFSRGPDLCLGYTDDALTAK-AFDDEGWYRT
Mb2405|M.bovis_AF2122/97 ELRIVNADGEPVGPGEEGELLVRGPYTLNGYFAAERDNERCFDPDGFYRS
Rv2384|M.tuberculosis_H37Rv ELRIVNADGEPVGPGEEGELLVRGPYTLNGYFAAERDNERCFDPDGFYRS
MAB_2247c|M.abscessus_ATCC_199 EVRIVDEDGADVPAGETGELLVRGPYTLNGYYRADADNMRSFTVDGFYRS
MUL_3145|M.ulcerans_Agy99 SVRLFDGD-RDVTASGRGQPGCRGPATSLGYLGGTDHEK-LFTRDGWMRM
MLBr_02661|M.leprae_Br4923 QIRIVGSDGQPLPPDTVGEVWLRGSTVVRGYLGDPAITAANFTH-GWLRT
*: **. ** *: *
MAV_1839|M.avium_104 GDRGYLRPDGRIVITGRTKDLIIRKGENIAPDEIENELLAHPLVDEVAVV
TH_3528|M.thermoresistible__bu GDSGHLRPDGRIVITGRVKDLIIRKGENISPEEIENELLAHPLIDQVAVI
Mflv_1784|M.gilvum_PYR-GCK GDLGRFTEDNHLVVTGRAKDIIIRNGENISPKEVEDILVTHPRIAEIAVV
Mvan_4966|M.vanbaalenii_PYR-1 GDLGHLTDDGYLVVTGRAKDLIIRNGENISPKEVEDILVTHPQIAEIAIV
MMAR_0303|M.marinum_M GDLGHWVDDEYLVVTGRAKDIIIRNGENISPKEIEDILITHPGIADVAIV
MSMEG_4774|M.smegmatis_MC2_155 GDVGFLDDDGYLTITDRTADLIIRGGENISALEVEEVLLGIPGVIEAIVV
Mb2405|M.bovis_AF2122/97 GDLVRRRDDGNLVVTGRVKDVICRAGETIAASDLEEQLLSHPAIFSAAAV
Rv2384|M.tuberculosis_H37Rv GDLVRRRDDGNLVVTGRVKDVICRAGETIAASDLEEQLLSHPAIFSAAAV
MAB_2247c|M.abscessus_ATCC_199 GDRVRALPGGYLEVTGRIKDVILRGGESIAALDLESHLQTHPAVYAAAAV
MUL_3145|M.ulcerans_Agy99 GDICEIDPDGYLTVTGRTSDFILRGGKNISAGQVEDAALTHPAVAIAAAV
MLBr_02661|M.leprae_Br4923 GDLGSLSVTGDLRIRGRIKELINRSGEKISPERVEGVLASHHNVMEVAVF
** : : .* :.* * *:.*:. :* : .
MAV_1839|M.avium_104 GQPDDLRGEMVCAVVRRSPRHRDVTLDELCAFLDQRGMMKQKWPERLFFV
TH_3528|M.thermoresistible__bu GHPDEERGELVCAVVRRSPGRRDLTLGEVCAFLDERGLMKQKWPERLVVV
Mflv_1784|M.gilvum_PYR-GCK GIPDPRTGERACAALVTTE-DPPPDIAELKDFLTDRGVARFKVPEQIHVV
Mvan_4966|M.vanbaalenii_PYR-1 GIPDLRTGERACAVIVAAD-SAAPDVAEIRDFLVARGVAKFKAPERVVVV
MMAR_0303|M.marinum_M GLPDDRTGERACAVIVAAA-DPPPGLPGVRAFLEGAGVARFKIPEQVVIW
MSMEG_4774|M.smegmatis_MC2_155 AAPDARLGERVAAVLRTRSGAPLPTLDDVRAHFERVGIARQKWPEELHHV
Mb2405|M.bovis_AF2122/97 GLPDQYLGEKICAAVVFAG--APITLAELNGYLDRRGVAAHTRPDQLVAM
Rv2384|M.tuberculosis_H37Rv GLPDQYLGEKICAAVVFAG--APITLAELNGYLDRRGVAAHTRPDQLVAM
MAB_2247c|M.abscessus_ATCC_199 GLPDQYLGEIVCAAIVFKG--KPVAAAELNQHLQARGAATHSRVDKLVAV
MUL_3145|M.ulcerans_Agy99 SMPDPVFGEKVCLYVELAD-SQRLDRTELVEHLLALGYSKELLPERLVVV
MLBr_02661|M.leprae_Br4923 GDPDKVYGETVTAVIVPRE-VIAPTPSELAVFCRDR-LAAFEVPTRFQEA
. ** ** : : . ..
MAV_1839|M.avium_104 D---EFPLTGLGKVAKAELVQQISGGRR---------
TH_3528|M.thermoresistible__bu D---EFPMTGLGKIAKRELVQQLTGGIR---------
Mflv_1784|M.gilvum_PYR-GCK G---ALPKNDAGKVLKHVLKAELTKES----------
Mvan_4966|M.vanbaalenii_PYR-1 D---ALPKNDAGKVLKHAIRAVLLEKPREGEP-----
MMAR_0303|M.marinum_M D---HLPKNHAGKIVKHRIKAALMKAEQ---------
MSMEG_4774|M.smegmatis_MC2_155 PDGQDFPRTASGKVQKYRVRQAVRNPAAVMDIATSAN
Mb2405|M.bovis_AF2122/97 P---ALPTTPIGKIDKRAIVRQLGIATGPVTTQRCH-
Rv2384|M.tuberculosis_H37Rv P---ALPTTPIGKIDKRAIVRQLGIATGPVTTQRCH-
MAB_2247c|M.abscessus_ATCC_199 S---ALPLTAVGKIDKRALLASL---TG---------
MUL_3145|M.ulcerans_Agy99 D---ELPRSSGGKIAKGQLRDDIRARMEAEHERS---
MLBr_02661|M.leprae_Br4923 S---ALPHTAKGSLDRRAVAEQFAHRG----------
:* . *.: : : .