For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TAGTIAGLLDECAAGRPDRPLLRDVDGETLTVSQVAGLASAAAGWLTDAGVRPGMTVAWQLPSHVNAAVA MLALARMPVVQAPVLHLYRCREVCAAVDVARADILLVDESTAANAAPGLPTIMVPADLVRRLQTSPATSA EYEALPSAAEPRWVYFTSGTTGRPKAVRHSDATLLSAARGYVAHLGVGSHPREVGTIAFPIAHIGGMVYL ATALFGGFPVVLIPKVSAADLPRVLAEHQVTVTGASTAFYQMLLAAQMAAPTTELLVPSLRMLIGGGAPC PPEVHKQVREHLRVPVVHAYGMTEAPMICVSEATDTDEQLANSAGRPIPGSQVRIGANGEIELRGANLTA GYLQHDQWADALTADGWFRSGDRGHLRPDGRIVVTGRTKDLIIRKGENVAPDEIENELLAHPLVDEIAVL GQPDELRGELVCAVVRRSPRHRDVTLDELCTFLDQRGLMKQKWPERLVLVDEFPLTGLGKVAKSELARQI AGGTR*
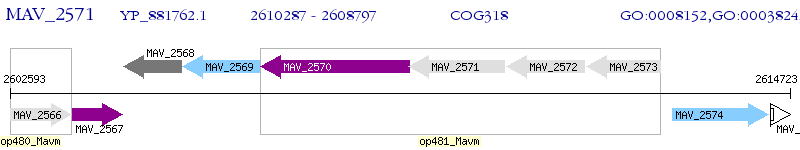
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2571 | - | - | 100% (496) | cyclohex-1-ene-1-carboxylate:CoA ligase |
| M. avium 104 | MAV_1839 | - | 0.0 | 71.34% (499) | cyclohex-1-ene-1-carboxylate:CoA ligase |
| M. avium 104 | MAV_2116 | - | 1e-82 | 38.31% (509) | AMP-binding enzyme, putative |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2405 | mbtA | 3e-43 | 30.39% (543) | bifunctional salicyl-AMP ligase/salicyl-S-arcp synthetase |
| M. gilvum PYR-GCK | Mflv_1784 | - | 4e-58 | 33.99% (509) | AMP-dependent synthetase and ligase |
| M. tuberculosis H37Rv | Rv2384 | mbtA | 3e-43 | 30.39% (543) | bifunctional salicyl-AMP ligase/salicyl-S-arcp synthetase |
| M. leprae Br4923 | MLBr_02661 | - | 5e-35 | 26.88% (532) | acyl-CoA synthetase |
| M. abscessus ATCC 19977 | MAB_2247c | - | 4e-45 | 30.10% (525) | bifunctional salicyl-AMP ligase/salicyl-S-arcp synthetase |
| M. marinum M | MMAR_0303 | - | 3e-50 | 31.43% (509) | acyl-CoA synthetase |
| M. smegmatis MC2 155 | MSMEG_4774 | - | 2e-51 | 32.28% (505) | AMP-dependent synthetase and ligase |
| M. thermoresistible (build 8) | TH_3528 | - | 0.0 | 70.12% (492) | PUTATIVE cyclohex-1-ene-1-carboxylate:CoA ligase |
| M. ulcerans Agy99 | MUL_3641 | mbtA | 3e-42 | 29.49% (529) | bifunctional enzyme MbtA: salicyl-AMP ligase (SAL-AMP |
| M. vanbaalenii PYR-1 | Mvan_4966 | - | 2e-58 | 34.27% (499) | AMP-dependent synthetase and ligase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2405|M.bovis_AF2122/97 -MPPKAADGRRPSPDGGLGGFVPFPADRAASYRAAGYWSGRTLDTVLSDA
Rv2384|M.tuberculosis_H37Rv -MPPKAADGRRPSPDGGLGGFVPFPADRAASYRAAGYWSGRTLDTVLSDA
MUL_3641|M.ulcerans_Agy99 -----------------MAGFIPYPPERVASYRAAGYWTGRSIGSLLRQA
MAB_2247c|M.abscessus_ATCC_199 MTQPRSTTTASSAATPALDGFVPFPSVRADEYRRAGYWTGNTVESLLRVS
MAV_2571|M.avium_104 -------------------------------------MTAGTIAGLLDEC
TH_3528|M.thermoresistible__bu -------------------------------------MTA-TIAGLLDDA
Mflv_1784|M.gilvum_PYR-GCK --------------------------------------MTGTLVEALRDA
Mvan_4966|M.vanbaalenii_PYR-1 --------------------------------------MT-TLADALRDA
MMAR_0303|M.marinum_M -----MGGPKVPQRRLIDGRWVRWDADRAAGAYAHRWWVRRTLADALREA
MSMEG_4774|M.smegmatis_MC2_155 --------------------MRDIPAELTKRYVEEGWWTPETLGQMLAEG
MLBr_02661|M.leprae_Br4923 --------------MKSESITSSRAPVEQIDARLTGQFESPSIASLVEAA
:: :
Mb2405|M.bovis_AF2122/97 ARRWPDRLAVADAGDRPGHGGLSYAELDQRADRAAAALHGLGITPGDRVL
Rv2384|M.tuberculosis_H37Rv ARRWPDRLAVADAGDRPGHGGLSYAELDQRADRAAAALHGLGITPGDRVL
MUL_3641|M.ulcerans_Agy99 AARWPDRVGVVDAG-----ASHTFCELDKLADRVAAGFVSLGIAPGDRVL
MAB_2247c|M.abscessus_ATCC_199 AARRPCHPAVIDEH-----RALSYRELDEAADNAAHAFLRLGIAPGDRVL
MAV_2571|M.avium_104 AAGRPDRPLLRDVD--GETLT--VSQVAGLASAAAGWLTDAGVRPGMTVA
TH_3528|M.thermoresistible__bu AARKPDQRMLIDVD--GGCLT--VAEVARLSSAATRWLADAGVQPGMTVA
Mflv_1784|M.gilvum_PYR-GCK ARDTPDRVLVRDDA--VALTC---AELLSAATGLAHALTRR-MPAGSVVS
Mvan_4966|M.vanbaalenii_PYR-1 ARDTPDRILLRDDE--ASLSS---AELLSAATNLAHALVRR-MPVGSVVS
MMAR_0303|M.marinum_M AQSTPQRVALVDAD--RQLDC---QELHRQAMALAQHLRSR-MPAGSVVS
MSMEG_4774|M.smegmatis_MC2_155 LAAGPDVVFCVHSE--VRPYTGTFGEVETLARRLAAGLRARGVGPGDVIA
MLBr_02661|M.leprae_Br4923 AIRNPSAPALVVTDN---RIVVSYRDLLRLVDDLTVQLALGGLLPGDRVA
* :: : : : * :
Mb2405|M.bovis_AF2122/97 LQLPNGCQFAVALFALLRAGAIPVMCLPGHRAAELGHFAAVSAATGLVVA
Rv2384|M.tuberculosis_H37Rv LQLPNGCQFAVALFALLRAGAIPVMCLPGHRAAELGHFAAVSAATGLVVA
MUL_3641|M.ulcerans_Agy99 LQLPNSCRFAVALFGLLRAGAIPVMCLPGHRLAELHHFAEVSGAVGLIVA
MAB_2247c|M.abscessus_ATCC_199 LQLPNRASFAVALFGLMRAGAIPVMCLPGHRAAELSHFIEVADAVGLVIP
MAV_2571|M.avium_104 WQLPSHVNAAVAMLALARMPVVQAPVLHLYRCREVCAAVDVARADILLVD
TH_3528|M.thermoresistible__bu WQLPAHSAAALLMLALARMPVVQAPVLHLYRQREVQAALDVAGADVLIVD
Mflv_1784|M.gilvum_PYR-GCK FTVPNWHEAAIIYLGATLAGMVVNPILPSLRERELSFILDDADSRMIFVP
Mvan_4966|M.vanbaalenii_PYR-1 FAVPNWHEAAVIYLGATLAGMVVNPILPSLRERELSFILADAGSRAIFIP
MMAR_0303|M.marinum_M FMLPNWHEAAVIYLASTLAAMVANPVLPSLREHELRFILEDAGSRMIFAP
MSMEG_4774|M.smegmatis_MC2_155 FQLPNWMEAAAAFWATSFLGAVTVPIVHFYGRKELAHILSTAKPKVFLTT
MLBr_02661|M.leprae_Br4923 LCAASNIEFVVGLLAASRAGLIVVPLDPALPVNEQCIRSQAAGVRVTLVD
. . . : * : .
Mb2405|M.bovis_AF2122/97 DVASGFDYRPMARELVADHPTLRHVIVDGDPG--PFVSWAQLCAQAGTGS
Rv2384|M.tuberculosis_H37Rv DVASGFDYRPMARELVADHPTLRHVIVDGDPG--PFVSWAQLCAQAGTGS
MUL_3641|M.ulcerans_Agy99 QHAGGFDYRSMAERLVAAHPQLRHVIIDGESG--PFLPWSSLSEGA---T
MAB_2247c|M.abscessus_ATCC_199 DSAGGFDYRALAVALSQRHPRLRTVVVEGDAG--PFASWTSLLKSVEPQG
MAV_2571|M.avium_104 ESTAANAAPGLPTIMVP-------------------ADLVRRLQTSPATS
TH_3528|M.thermoresistible__bu ASTAANAPDDTSVITLP-------------------DDFVDVLRAMPAEP
Mflv_1784|M.gilvum_PYR-GCK ESFGGHDYAAMLDRVAPGLPSPPEVVVVRGE---GRVTLADLLA-EPADG
Mvan_4966|M.vanbaalenii_PYR-1 QTFGGHDYAAMLDRVVAGLPSPPEVVVVRGEP--RRPALTDLVS-EPRDG
MMAR_0303|M.marinum_M ADFRGHDYTSMLRRVAAQLPSRPEVIEVRGQCPDGVSTFQSLLVGRETAA
MSMEG_4774|M.smegmatis_MC2_155 ERFGRMSFQPDLCAQVP-------IVGLVGES-----SFTDLLAERPMSG
MLBr_02661|M.leprae_Br4923 SLALEGVSDQRAATMRYWPIAVSYGSVTGASE---GSLLVHLDDTAALHP
Mb2405|M.bovis_AF2122/97 PAPPA--DPGSPALLLVSGGTTGMPKLIPRTHDDYVFNATASAALCRLS-
Rv2384|M.tuberculosis_H37Rv PAPPA--DPGSPALLLVSGGTTGMPKLIPRTHDDYVFNATASAALCRLS-
MUL_3641|M.ulcerans_Agy99 PDIDV--DTTSPALLLVSGGTTGTPKLIPRTHDDYVYNATASAQLCRLT-
MAB_2247c|M.abscessus_ATCC_199 PLPTV--QTDTPALLLVSGGTTAAPKLIPRTHEDYVYNATQAARVCGLT-
MAV_2571|M.avium_104 AEYEALPSAAEPRWVYFTSGTTGRPKAVRHSDATLLSAARGYVAHLGVGS
TH_3528|M.thermoresistible__bu VLVED-PRPDDVRWIYFTSGTTGRPKGVRHTDSTLLAAARGFTAHLGLGT
Mflv_1784|M.gilvum_PYR-GCK VLPAP--RPTDTRMILYTSGTTGRPKGVLHTHESLAALIAQLGRFWRI--
Mvan_4966|M.vanbaalenii_PYR-1 TLPDL--DPAATRMILYTSGTTGRPKGVLHSHKSLGALIAQLGRYWRI--
MMAR_0303|M.marinum_M PLPTA--DPDAVRMVLYTSGTTGRPKGVLHTHNSIHALICQIRDHWMV--
MSMEG_4774|M.smegmatis_MC2_155 TLDT---DPAAPALIAFTSGTTRDPKGVIHSHQTLSCETRQLLANYPP--
MLBr_02661|M.leprae_Br4923 VTSTPDGLRHDDAMIMFTGGTTGLPKMVPWTDGNIAGSVHAIITAYQLG-
: :.*** ** : :.
Mb2405|M.bovis_AF2122/97 -ADDVYLVVLAAGHNFPLACPGLLGAMTVGATAVFAP--DPSPEAAFAAI
Rv2384|M.tuberculosis_H37Rv -ADDVYLVVLAAGHNFPLACPGLLGAMTVGATAVFAP--DPSPEAAFAAI
MUL_3641|M.ulcerans_Agy99 -ADDVYLVSLPAAHNFPLACPGILGSMAVGATIVFST--DPSPEAAFTLI
MAB_2247c|M.abscessus_ATCC_199 -QDDVYLVALPAGHNFPLACPGLLGAISTGATTVFLS--DPSPESAFATI
MAV_2571|M.avium_104 HPREVGTIAFPIAHIGGMVY--LATALFGGFPVVLIP--KVSAADLPRVL
TH_3528|M.thermoresistible__bu HPQEVSTIAFPVAHIGGIVY--LSCALLAGFPVLLIP--KVEADELLRLL
Mflv_1784|M.gilvum_PYR-GCK DRGDTFLVPSPIAHIGGSIYA-FECPLLLGTEAVLMQ--RWDPDAAVGLM
Mvan_4966|M.vanbaalenii_PYR-1 DPGDTFLVPSPIAHIGGSIYA-FECPLLLGTQAVLMQ--RWDPDAAVALM
MMAR_0303|M.marinum_M EPGDGFLVASPIAHIGGSIYA-FECPLLLGTTAVLLD--RWDADRAVQLM
MSMEG_4774|M.smegmatis_MC2_155 NRG-RQLTATPVGHFIGMVGA-FLIPVLEGAPVDLCD--VWDPAKVLRLM
MLBr_02661|M.leprae_Br4923 -PQDATVVVMPLYHGHGLIAA-LLSTLASGGVVLLPARARFSARTFWDDI
. * : .: * : .. :
Mb2405|M.bovis_AF2122/97 ERHGVTVTALVPALAKLWAQSCEWEPVTP---KSLRLLQVGGSKLEPEDA
Rv2384|M.tuberculosis_H37Rv ERHGVTVTALVPALAKLWAQSCEWEPVTP---KSLRLLQVGGSKLEPEDA
MUL_3641|M.ulcerans_Agy99 ERHGVTVTALVPALAKLWTQACDWEPVIP---KSLRLLQVGGSRLEPEDA
MAB_2247c|M.abscessus_ATCC_199 ARHRVSVTAVVPSLAQLWAQATAWEPVLP---ETLRLLQVGGAKLGADDA
MAV_2571|M.avium_104 AEHQVTVTGASTAFYQMLLAAQMAAPTTELLVPSLRMLIGGGAPCPPEVH
TH_3528|M.thermoresistible__bu AEYRVTVTGGSTAFYQMLLSAQLASGQNEPLVPTLRMLIGGGAPCPPEVH
Mflv_1784|M.gilvum_PYR-GCK LEHRVTHMAGATPFLVGLLAAAQRAGTR---LPDLKVFICGGASVPPTLI
Mvan_4966|M.vanbaalenii_PYR-1 LRHRCTHMAGATPFLDGLLAAAERAGTR---LPDLKVFICGGASVPPSLI
MMAR_0303|M.marinum_M TSKRCTHMAGATPFLEQLLAAAQRAGTR---LPDLKFFVCGGASVSPSLI
MSMEG_4774|M.smegmatis_MC2_155 ESDGLSIGGGPPYFVTSLLDHPDFADRH---LARIRTVGLGGSTVPAAVT
MLBr_02661|M.leprae_Br4923 DAVAATWYTAAPAIHRILLELASTQSFRSK-RAKLRFIRSCSAPLTQETA
: . : :: . .:
Mb2405|M.bovis_AF2122/97 RRVRTALT-PGLQQVFGMAEG-----LLNFTRIGDPPEVVEHTQGRPLCP
Rv2384|M.tuberculosis_H37Rv RRVRTALT-PGLQQVFGMAEG-----LLNFTRIGDPPEVVEHTQGRPLCP
MUL_3641|M.ulcerans_Agy99 RQAREALT-PGLQQVFGMAEG-----LLNFTRLEDPPDLLEHTQGRPLCS
MAB_2247c|M.abscessus_ATCC_199 RAVRESLT-QGLQQVFGMAEG-----LLCLTQPGDPTDILDNSQGRPMCD
MAV_2571|M.avium_104 KQVREHLR-VPVVHAYGMTEAP----MICVSEATDTDEQLANSAGRPIPG
TH_3528|M.thermoresistible__bu RKVRAHMG-IPIVHAYGMTEAP----MIGVSEAADTEEQQSNSSGRPIPG
Mflv_1784|M.gilvum_PYR-GCK RSAAAYFDKAAVSRVYGSTEVP----VTTVG---ALDDIDRAADTDGRPG
Mvan_4966|M.vanbaalenii_PYR-1 RRAADYFDKAAVSRVYGSTEVP----VTTVG---SLDDVDRAADTDGRPG
MMAR_0303|M.marinum_M RCATDYFDKAVVTRVYGSTEVP----VTTIG---APDSSAHAAATDGRVG
MSMEG_4774|M.smegmatis_MC2_155 RRLADLG--IFVFRSYGSTEHP----SITGSSPSAPEGKRLYTDGNPRPG
MLBr_02661|M.leprae_Br4923 QALREEFL-APVICAFGMTEATHQVTTTNIKWFGQGENPTVTNGLVGQST
: : :* :*
Mb2405|M.bovis_AF2122/97 ADELRIVNADGEPVGPGEEGELLVRGPYTLNGYFAAERDNERCFDPDGFY
Rv2384|M.tuberculosis_H37Rv ADELRIVNADGEPVGPGEEGELLVRGPYTLNGYFAAERDNERCFDPDGFY
MUL_3641|M.ulcerans_Agy99 ADELRIVDGAGAPVAPGAEGELLVRGPYTINGYFCADRDNERCFDPAGFY
MAB_2247c|M.abscessus_ATCC_199 ADEVRIVDEDGADVPAGETGELLVRGPYTLNGYYRADADNMRSFTVDGFY
MAV_2571|M.avium_104 SQVRIGAN-----------GEIELRGANLTAGYLQHDQWAD-ALTADGWF
TH_3528|M.thermoresistible__bu SEIRIAAN-----------GEIXLRGSNVTTGYVX-DQWSG-STTADGWF
Mflv_1784|M.gilvum_PYR-GCK IAEVRIVD-----------GEIRARGPQMFTGYLHPEDDRE-SFDDAGYF
Mvan_4966|M.vanbaalenii_PYR-1 IADVTLVD-----------GEIRARGPQMLTGYLHPEDNSE-SFDAEGFF
MMAR_0303|M.marinum_M AADVKLVA-----------GEIRVRGPQMLVGYRHPEDEAG-SFDAQGYF
MSMEG_4774|M.smegmatis_MC2_155 VEIRLGPD-----------GEIFSRGPDLCLGYTDDALTAK-AFDDEGWY
MLBr_02661|M.leprae_Br4923 GVQIRIVGSDGQPLPPDTVGEVWLRGSTVVRGYLGDPAITAANFTH-GWL
**: **. ** *:
Mb2405|M.bovis_AF2122/97 RSGDLVRRRDDGNLVVTGRVKDVICRAGETIAASDLEEQLLSHPAIFSAA
Rv2384|M.tuberculosis_H37Rv RSGDLVRRRDDGNLVVTGRVKDVICRAGETIAASDLEEQLLSHPAIFSAA
MUL_3641|M.ulcerans_Agy99 RSGDLVRLRDDGYLQVTGRVKDVICRCGETISAQDLEEQLLSHPAIRSAA
MAB_2247c|M.abscessus_ATCC_199 RSGDRVRALPGGYLEVTGRIKDVILRGGESIAALDLESHLQTHPAVYAAA
MAV_2571|M.avium_104 RSGDRGHLRPDGRIVVTGRTKDLIIRKGENVAPDEIENELLAHPLVDEIA
TH_3528|M.thermoresistible__bu RTGDSGHLRPDGRIVITGRVKDLIIRKGENISPEEIENELLAHPLIDQVA
Mflv_1784|M.gilvum_PYR-GCK RTGDLGRFTEDNHLVVTGRAKDIIIRNGENISPKEVEDILVTHPRIAEIA
Mvan_4966|M.vanbaalenii_PYR-1 RTGDLGHLTDDGYLVVTGRAKDLIIRNGENISPKEVEDILVTHPQIAEIA
MMAR_0303|M.marinum_M RTGDLGHWVDDEYLVVTGRAKDIIIRNGENISPKEIEDILITHPGIADVA
MSMEG_4774|M.smegmatis_MC2_155 RTGDVGFLDDDGYLTITDRTADLIIRGGENISALEVEEVLLGIPGVIEAI
MLBr_02661|M.leprae_Br4923 RTGDLGSLSVTGDLRIRGRIKELINRSGEKISPERVEGVLASHHNVMEVA
*:** : : .* ::* * **.::. :* * :
Mb2405|M.bovis_AF2122/97 AVGLPDQYLGEKICAAVVFAG---APITLAELNGYLDRRGVAAHTRPDQL
Rv2384|M.tuberculosis_H37Rv AVGLPDQYLGEKICAAVVFAG---APITLAELNGYLDRRGVAAHTRPDQL
MUL_3641|M.ulcerans_Agy99 AVPLPDPDMGELICAAIVFAGPPQTPQTLSGLNDYLEQRGVAAHIRIDRL
MAB_2247c|M.abscessus_ATCC_199 AVGLPDQYLGEIVCAAIVFKG---KPVAAAELNQHLQARGAATHSRVDKL
MAV_2571|M.avium_104 VLGQPDELRGELVCAVVRRSPR-HRDVTLDELCTFLDQRGLMKQKWPERL
TH_3528|M.thermoresistible__bu VIGHPDEERGELVCAVVRRSPG-RRDLTLGEVCAFLDERGLMKQKWPERL
Mflv_1784|M.gilvum_PYR-GCK VVGIPDPRTGERACAALVTTE--DPPPDIAELKDFLTDRGVARFKVPEQI
Mvan_4966|M.vanbaalenii_PYR-1 IVGIPDLRTGERACAVIVAAD--SAAPDVAEIRDFLVARGVAKFKAPERV
MMAR_0303|M.marinum_M IVGLPDDRTGERACAVIVAAA--DPPPGLPGVRAFLEGAGVARFKIPEQV
MSMEG_4774|M.smegmatis_MC2_155 VVAAPDARLGERVAAVLRTRSG-APLPTLDDVRAHFERVGIARQKWPEEL
MLBr_02661|M.leprae_Br4923 VFGDPDKVYGETVTAVIVPREV--IAPTPSELAVFCRDR-LAAFEVPTRF
. ** ** *.: : . ..
Mb2405|M.bovis_AF2122/97 VAMP---ALPTTPIGKIDKRAIVRQLGIATGPVTTQRCH-
Rv2384|M.tuberculosis_H37Rv VAMP---ALPTTPIGKIDKRAIVRQLGIATGPVTTQRCH-
MUL_3641|M.ulcerans_Agy99 VAMP---SLPTTPVGKIDKKAIASQILTTTAT--------
MAB_2247c|M.abscessus_ATCC_199 VAVS---ALPLTAVGKIDKRALLASLTG------------
MAV_2571|M.avium_104 VLVD---EFPLTGLGKVAKSELARQIAGGTR---------
TH_3528|M.thermoresistible__bu VVVD---EFPMTGLGKIAKRELVQQLTGGIR---------
Mflv_1784|M.gilvum_PYR-GCK HVVG---ALPKNDAGKVLKHVLKAELTKES----------
Mvan_4966|M.vanbaalenii_PYR-1 VVVD---ALPKNDAGKVLKHAIRAVLLEKPREGEP-----
MMAR_0303|M.marinum_M VIWD---HLPKNHAGKIVKHRIKAALMKAEQ---------
MSMEG_4774|M.smegmatis_MC2_155 HHVPDGQDFPRTASGKVQKYRVRQAVRNPAAVMDIATSAN
MLBr_02661|M.leprae_Br4923 QEAS---ALPHTAKGSLDRRAVAEQFAHRG----------
:* . *.: : : .