For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPPKAADGRRPSPDGGLGGFVPFPADRAASYRAAGYWSGRTLDTVLSDAARRWPDRLAVADAGDRPGHGG LSYAELDQRADRAAAALHGLGITPGDRVLLQLPNGCQFAVALFALLRAGAIPVMCLPGHRAAELGHFAAV SAATGLVVADVASGFDYRPMARELVADHPTLRHVIVDGDPGPFVSWAQLCAQAGTGSPAPPADPGSPALL LVSGGTTGMPKLIPRTHDDYVFNATASAALCRLSADDVYLVVLAAGHNFPLACPGLLGAMTVGATAVFAP DPSPEAAFAAIERHGVTVTALVPALAKLWAQSCEWEPVTPKSLRLLQVGGSKLEPEDARRVRTALTPGLQ QVFGMAEGLLNFTRIGDPPEVVEHTQGRPLCPADELRIVNADGEPVGPGEEGELLVRGPYTLNGYFAAER DNERCFDPDGFYRSGDLVRRRDDGNLVVTGRVKDVICRAGETIAASDLEEQLLSHPAIFSAAAVGLPDQY LGEKICAAVVFAGAPITLAELNGYLDRRGVAAHTRPDQLVAMPALPTTPIGKIDKRAIVRQLGIATGPVT TQRCH
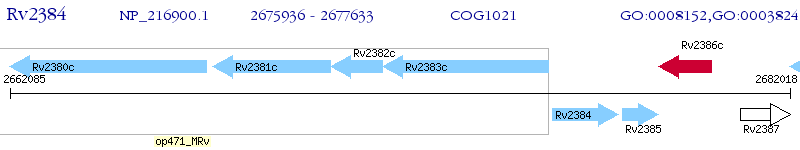
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2384 | mbtA | - | 100% (565) | BIFUNCTIONAL ENZYME MBTA: SALICYL-AMP LIGASE (SAL-AMP LIGASE) + SALICYL-S-ArCP SYNTHETASE |
| M. tuberculosis H37Rv | Rv0119 | fadD7 | 8e-42 | 29.03% (534) | acyl-CoA synthetase |
| M. tuberculosis H37Rv | Rv3561 | fadD3 | 6e-38 | 29.10% (536) | acyl-CoA synthetase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2405 | mbtA | 0.0 | 100.00% (565) | bifunctional salicyl-AMP ligase/salicyl-S-arcp synthetase |
| M. gilvum PYR-GCK | Mflv_2691 | - | 0.0 | 65.06% (538) | AMP-dependent synthetase and ligase |
| M. leprae Br4923 | MLBr_02661 | - | 2e-33 | 26.61% (545) | acyl-CoA synthetase |
| M. abscessus ATCC 19977 | MAB_2247c | - | 0.0 | 61.64% (550) | bifunctional salicyl-AMP ligase/salicyl-S-arcp synthetase |
| M. marinum M | MMAR_3698 | mbtA | 0.0 | 72.07% (555) | bifunctional enzyme MbtA: salicyl-AMP ligase (SAL-AMP |
| M. avium 104 | MAV_2008 | - | 0.0 | 78.33% (540) | 2,3-dihydroxybenzoate-AMP ligase |
| M. smegmatis MC2 155 | MSMEG_4516 | - | 0.0 | 69.19% (555) | 2,3-dihydroxybenzoate-AMP ligase |
| M. thermoresistible (build 8) | TH_2907 | - | 0.0 | 64.58% (542) | PUTATIVE AMP-dependent synthetase and ligase |
| M. ulcerans Agy99 | MUL_3641 | mbtA | 0.0 | 72.91% (539) | bifunctional enzyme MbtA: salicyl-AMP ligase (SAL-AMP |
| M. vanbaalenii PYR-1 | Mvan_3850 | - | 0.0 | 63.57% (538) | AMP-dependent synthetase and ligase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2691|M.gilvum_PYR-GCK ---MTTGFPIEYGHGLTNGDLKHGFVPFPEDRAEAYRRAGYWTGRPLDSI
Mvan_3850|M.vanbaalenii_PYR-1 ---MNTGLPADQGN-----DLTRGFVPFPPDRAEVYRRAGYWTGRPLDSI
TH_2907|M.thermoresistible__bu ---MTTEFQPVESDLTS--DFRTGFVPFPPDRVEEYRRAGYWTGRRLDSI
Rv2384|M.tuberculosis_H37Rv -----MPPKAADGRRPSPDGGLGGFVPFPADRAASYRAAGYWSGRTLDTV
Mb2405|M.bovis_AF2122/97 -----MPPKAADGRRPSPDGGLGGFVPFPADRAASYRAAGYWSGRTLDTV
MAV_2008|M.avium_104 -----MSPNTT-----PPAGVLDGFVPFPAERAAAYRAAGLWTGRALDTI
MMAR_3698|M.marinum_M -----MPVTSA---QPPRPGPVAGFIPYPPERVASYRAAGYWTGRSIGSL
MUL_3641|M.ulcerans_Agy99 ---------------------MAGFIPYPPERVASYRAAGYWTGRSIGSL
MSMEG_4516|M.smegmatis_MC2_155 MTLTTPHPRPEQSESAAQSSLLAGFTPFPAERAQAYRAAGYWRDQLLDSV
MAB_2247c|M.abscessus_ATCC_199 ----MTQPRSTTTASSAATPALDGFVPFPSVRADEYRRAGYWTGNTVESL
MLBr_02661|M.leprae_Br4923 ------------------MKSESITSSRAPVEQIDARLTGQFESPSIASL
. . . * :* : . : ::
Mflv_2691|M.gilvum_PYR-GCK LTDAASRWPDKPAIIDAT-----RSYTFAELDAAADRAAAGLADLGIRPG
Mvan_3850|M.vanbaalenii_PYR-1 LTEAAATWPDKPAVIDSA-----QTYTFRELDAAADRVAAGLAALGIRPG
TH_2907|M.thermoresistible__bu LSDAAAAQPDKAAVIDPD-----RSHTFAELDALADRVAAGLADLGIAPG
Rv2384|M.tuberculosis_H37Rv LSDAARRWPDRLAVADAGDRPGHGGLSYAELDQRADRAAAALHGLGITPG
Mb2405|M.bovis_AF2122/97 LSDAARRWPDRLAVADAGDRPGHGGLSYAELDQRADRAAAALHGLGITPG
MAV_2008|M.avium_104 LTDAARRWPDRTAVLDAS---GGTGFSYAGLDEQANRAAAGLADAGIAPG
MMAR_3698|M.marinum_M LRQAAARWPDRVGVVDAG-----ASHTFCELDKLADRAAAGFVSLGIAPG
MUL_3641|M.ulcerans_Agy99 LRQAAARWPDRVGVVDAG-----ASHTFCELDKLADRVAAGFVSLGIAPG
MSMEG_4516|M.smegmatis_MC2_155 LRTAARTWPDHIAVIDAD-----HRHTYAELDRLADRAAAGIAGLGIRPG
MAB_2247c|M.abscessus_ATCC_199 LRVSAARRPCHPAVIDEH-----RALSYRELDEAADNAAHAFLRLGIAPG
MLBr_02661|M.leprae_Br4923 VEAAAIRNPSAPALVVTDN---RIVVSYRDLLRLVDDLTVQLALGGLLPG
: :* * .: :: * .: : : *: **
Mflv_2691|M.gilvum_PYR-GCK ERVLLQLPNSCQFAVALFALLRAGVVPVMCLTGHRTAELGHFADVSGAVA
Mvan_3850|M.vanbaalenii_PYR-1 DRVLLQLPNSCQFAIALFGLLRAAAVPVMCLPGHRTAELGHFAEVSGAVA
TH_2907|M.thermoresistible__bu DRVLLQMPNDTRFAVAFFGLLRAGAIPVMCLPGHRSAEMSHFAEVSGAVG
Rv2384|M.tuberculosis_H37Rv DRVLLQLPNGCQFAVALFALLRAGAIPVMCLPGHRAAELGHFAAVSAATG
Mb2405|M.bovis_AF2122/97 DRVLLQLPNGCQFAVALFALLRAGAIPVMCLPGHRAAELGHFAAVSAATG
MAV_2008|M.avium_104 DRVLLQLPNGCQFAVALFALLRAGAIPVMCLPGHRAAELGHFAALSQATA
MMAR_3698|M.marinum_M DRVLLQLPNSCRFAVALFGLLRAGAIPVMCLPGHRLAELHHFAEVSGAVG
MUL_3641|M.ulcerans_Agy99 DRVLLQLPNSCRFAVALFGLLRAGAIPVMCLPGHRLAELHHFAEVSGAVG
MSMEG_4516|M.smegmatis_MC2_155 DRVLVQLPNTAEFAVALFGLLRAGAVPVMCLPGHRLAELTHFAEVSSAVA
MAB_2247c|M.abscessus_ATCC_199 DRVLLQLPNRASFAVALFGLMRAGAIPVMCLPGHRAAELSHFIEVADAVG
MLBr_02661|M.leprae_Br4923 DRVALCAASNIEFVVGLLAASRAGLIVVPLDPALPVNEQCIRSQAAGVRV
:** : .. *.:.::. **. : * .. * : .
Mflv_2691|M.gilvum_PYR-GCK VIVPDRAGGFDYRDMAASLAAQRPALRHVIVDGEPGD----QVSWQSILD
Mvan_3850|M.vanbaalenii_PYR-1 LIVPDEAGGFDYRDMAGRLAAEHPSLQHVIVDGENGP----FVAWSSIRD
TH_2907|M.thermoresistible__bu LIIPERANGFDYRELARALTASRPRLRHVIVDGATDPSGDGFVPWSALAE
Rv2384|M.tuberculosis_H37Rv LVVADVASGFDYRPMARELVADHPTLRHVIVDGDPGP----FVSWAQLCA
Mb2405|M.bovis_AF2122/97 LVVADVASGFDYRPMARELVADHPTLRHVIVDGDPGP----FVSWAQLCA
MAV_2008|M.avium_104 LLIADTAAGFDYRTMAAGLVEEHEALAHVIVDGDPGP----FLSWAQLCE
MMAR_3698|M.marinum_M LIVAQHAGGFDYRSMAERLVAAHPQLRHVIIDGESGP----FLPWSSLSE
MUL_3641|M.ulcerans_Agy99 LIVAQHAGGFDYRSMAERLVAAHPQLRHVIIDGESGP----FLPWSSLSE
MSMEG_4516|M.smegmatis_MC2_155 LVVADTAGGFDHRDLARELVRSHPDVRHVLVDGDAAE----FLSWAEVTR
MAB_2247c|M.abscessus_ATCC_199 LVIPDSAGGFDYRALAVALSQRHPRLRTVVVEGDAGP----FASWTSLLK
MLBr_02661|M.leprae_Br4923 TLVDSLALEGVSDQRAATMRYWPIAVSYGSVTGASEGS---LLVHLDDTA
:: . * * : : : *
Mflv_2691|M.gilvum_PYR-GCK FAGPRLGRTPVDPSLPALLLVSGGTTGLPKLIARTHDDYVYNAHACAQAC
Mvan_3850|M.vanbaalenii_PYR-1 FDGPAVERAAVDPGLPALLLVSGGTTGLPKLIARTHDDYVYNAVASAQAC
TH_2907|M.thermoresistible__bu FAGPAVPRPPVDTTAPALLLVSGGTTGLPKLIPRTHDDYVYTAVASARAY
Rv2384|M.tuberculosis_H37Rv QAGTGSPAPPADPGSPALLLVSGGTTGMPKLIPRTHDDYVFNATASAALC
Mb2405|M.bovis_AF2122/97 QAGTGSPAPPADPGSPALLLVSGGTTGMPKLIPRTHDDYVFNATASAALC
MAV_2008|M.avium_104 RAPAGRPATPVDPGSPALLLVSGGTTGTPKLIPRTHDDYVFNANASAELC
MMAR_3698|M.marinum_M GA---TPDIDVDTTSPALLLVSGGTTGTPKLIPRTHDDYVYNATASAQLC
MUL_3641|M.ulcerans_Agy99 GA---TPDIDVDTTSPALLLVSGGTTGTPKLIPRTHDDYVYNATASAQLC
MSMEG_4516|M.smegmatis_MC2_155 AAPGPVPEIAPDPAAPALLLVSGGTTGAPKLIPRTHQDYVYNATASAELC
MAB_2247c|M.abscessus_ATCC_199 SVEPQGPLPTVQTDTPALLLVSGGTTAAPKLIPRTHEDYVYNATQAARVC
MLBr_02661|M.leprae_Br4923 ALHPVTSTPDGLRHDDAMIMFTGGTTGLPKMVPWTDGNIAGSVHAIITAY
*:::.:****. **::. *. : . ..
Mflv_2691|M.gilvum_PYR-GCK EMTGEDAYLVALPAGHNFPLACPGLLGSMTVGAPTVFTADP--SPENAFA
Mvan_3850|M.vanbaalenii_PYR-1 EMTAEDAYLVALPAGHNFPLACPGMLGSMTVGAPTVFTADP--SPENAFA
TH_2907|M.thermoresistible__bu EMTSDDVYLVVLPAAHNFPLACPGMLGAMTVGASTVFTADP--SPEAAFP
Rv2384|M.tuberculosis_H37Rv RLSADDVYLVVLAAGHNFPLACPGLLGAMTVGATAVFAPDP--SPEAAFA
Mb2405|M.bovis_AF2122/97 RLSADDVYLVVLAAGHNFPLACPGLLGAMTVGATAVFAPDP--SPEAAFA
MAV_2008|M.avium_104 GLTRDDVYLAVLSAGHNFPLACPGLLGAMTVGATTVFGTDP--SPEAAFA
MMAR_3698|M.marinum_M RLTADDVYLVSLPAAHNFPLACPGILGSMAVGATIVFSTDP--SPEAAFT
MUL_3641|M.ulcerans_Agy99 RLTADDVYLVSLPAAHNFPLACPGILGSMAVGATIVFSTDP--SPEAAFT
MSMEG_4516|M.smegmatis_MC2_155 RLTADDVYLVALPAAHNFPLACPGLLGAMTVGATTVFTTDP--SPEAAFA
MAB_2247c|M.abscessus_ATCC_199 GLTQDDVYLVALPAGHNFPLACPGLLGAISTGATTVFLSDP--SPESAFA
MLBr_02661|M.leprae_Br4923 QLGPQDATVVVMPLYHGHGLIAA-LLSTLASGGVVLLPARARFSARTFWD
: :*. :. :. *.. * .. :*.::: *. :: . . *.. :
Mflv_2691|M.gilvum_PYR-GCK LIDKHRITVTGLVNALAKLWAQACDWEPVLPT--SLRFVQVGGSRMSPEE
Mvan_3850|M.vanbaalenii_PYR-1 LIDKFRITVTGLVNALAKLWAQACEWEPVLPT--SLRFVQVGGSRMSPEE
TH_2907|M.thermoresistible__bu LINRHRVTVTGLVNALAKLWAQACEWEPELPE--SLRVVQVGGSRMSPQE
Rv2384|M.tuberculosis_H37Rv AIERHGVTVTALVPALAKLWAQSCEWEPVTPK--SLRLLQVGGSKLEPED
Mb2405|M.bovis_AF2122/97 AIERHGVTVTALVPALAKLWAQSCEWEPVTPK--SLRLLQVGGSKLEPED
MAV_2008|M.avium_104 TIARHGVTVTALVPALAKLWAHACEWEDNPPK--SLRLLQVGGAKLEADD
MMAR_3698|M.marinum_M LIERHGVTVTALVPALAKLWTQACDWEPVTPK--SLRLLQVGGSRLEPED
MUL_3641|M.ulcerans_Agy99 LIERHGVTVTALVPALAKLWTQACDWEPVIPK--SLRLLQVGGSRLEPED
MSMEG_4516|M.smegmatis_MC2_155 AIDEHGVTATALVPALAKLWAQACAWEPLAPK--TLRLLQVGGAKLAAPD
MAB_2247c|M.abscessus_ATCC_199 TIARHRVSVTAVVPSLAQLWAQATAWEPVLPE--TLRLLQVGGAKLGADD
MLBr_02661|M.leprae_Br4923 DIDAVAATWYTAAPAIHRILLELASTQSFRSKRAKLRFIRSCSAPLTQET
* : . :: :: . : . .**.:: .: :
Mflv_2691|M.gilvum_PYR-GCK ARFILDRLTPGMSQIFGMAEGMLNFTRP-----GDPEDVVVHTQGRPMSP
Mvan_3850|M.vanbaalenii_PYR-1 ARFILERLTPGMSQIFGMAEGMLNFTRP-----GDPEDVVVHTQGRPMSP
TH_2907|M.thermoresistible__bu AQFILDRLTPGMSQIFGMAEGMLNFTRP-----GDPIDVVVHTQGRPMSP
Rv2384|M.tuberculosis_H37Rv ARRVRTALTPGLQQVFGMAEGLLNFTRI-----GDPPEVVEHTQGRPLCP
Mb2405|M.bovis_AF2122/97 ARRVRTALTPGLQQVFGMAEGLLNFTRI-----GDPPEVVEHTQGRPLCP
MAV_2008|M.avium_104 ARVIRSALTPGLQQVFGMAEGLLNYTRL-----DDPPEITEHTQGRPLST
MMAR_3698|M.marinum_M ARQAREALTPGLQQVFGMAEGLLNFTRL-----EDPPDLLEHTQGRPLCS
MUL_3641|M.ulcerans_Agy99 ARQAREALTPGLQQVFGMAEGLLNFTRL-----EDPPDLLEHTQGRPLCS
MSMEG_4516|M.smegmatis_MC2_155 AALVRGALTPGLQQVFGMAEGLLNYTRI-----GDPPEVLENTQGRPLSP
MAB_2247c|M.abscessus_ATCC_199 ARAVRESLTQGLQQVFGMAEGLLCLTQP-----GDPTDILDNSQGRPMCD
MLBr_02661|M.leprae_Br4923 AQALREEFLAPVICAFGMTEATHQVTTTNIKWFGQGENPTVTNGLVGQST
* : : ***:*. * : : . .
Mflv_2691|M.gilvum_PYR-GCK HDEMRVVDEDGVEVAPGEEGELLVRGPYTLNGYYRADDANARSFTPDGFY
Mvan_3850|M.vanbaalenii_PYR-1 HDEVRVVDESGDGVPPGEEGELLVRGPYTLYGYYRDDDANARSFSPDGFY
TH_2907|M.thermoresistible__bu HDEMRVVDESGAEVAPGEEGELLVRGPNTLNGYYRDDEANARSFSPDGFY
Rv2384|M.tuberculosis_H37Rv ADELRIVNADGEPVGPGEEGELLVRGPYTLNGYFAAERDNERCFDPDGFY
Mb2405|M.bovis_AF2122/97 ADELRIVNADGEPVGPGEEGELLVRGPYTLNGYFAAERDNERCFDPDGFY
MAV_2008|M.avium_104 ADEVRVVDAAGNPVAPGQEGELLVRGPYTFNGYFRAERDNQRCFDAQGFF
MMAR_3698|M.marinum_M ADELRIVDGAGAPVAPGAEGELLVRGPYTINGYFCADRDNERCFDPAGFY
MUL_3641|M.ulcerans_Agy99 ADELRIVDGAGAPVAPGAEGELLVRGPYTINGYFCADRDNERCFDPAGFY
MSMEG_4516|M.smegmatis_MC2_155 DDEIRIVDEVGNEVPPGAEGELLVRGPYTLNGYFNAEAANERSFSPDGFY
MAB_2247c|M.abscessus_ATCC_199 ADEVRIVDEDGADVPAGETGELLVRGPYTLNGYYRADADNMRSFTVDGFY
MLBr_02661|M.leprae_Br4923 GVQIRIVGSDGQPLPPDTVGEVWLRGSTVVRGYLGDPAITAANFT-HGWL
::*:*. * : .. **: :**. .. ** . * *:
Mflv_2691|M.gilvum_PYR-GCK RTGDRVRIFTDGPRAGYVEVTGRVKDVIHRGGETVSATDLEDHLHTHPSV
Mvan_3850|M.vanbaalenii_PYR-1 RTGDRVRIFADGPRAGYVEVVGRIKDVIHRGGETVSATDLEDHLHTHPAI
TH_2907|M.thermoresistible__bu RTGDRVRIFADGPRAGHVEVTGRVKDVIHRGGETVSATDLEDHLLTHPAI
Rv2384|M.tuberculosis_H37Rv RSGDLVRRRDD----GNLVVTGRVKDVICRAGETIAASDLEEQLLSHPAI
Mb2405|M.bovis_AF2122/97 RSGDLVRRRDD----GNLVVTGRVKDVICRAGETIAASDLEEQLLSHPAI
MAV_2008|M.avium_104 RSGDLVRVRDD----GYLVVTGRVKDVICRGGETISAADLEEQMLSHPAI
MMAR_3698|M.marinum_M RSGDLVRLRDD----GYLQVTGRVKDVICRCGETISAQDLEEQLLSHPAI
MUL_3641|M.ulcerans_Agy99 RSGDLVRLRDD----GYLQVTGRVKDVICRCGETISAQDLEEQLLSHPAI
MSMEG_4516|M.smegmatis_MC2_155 RSGDRVRRFADGPLAGYLEVTGRIKDVIVRGGENVSALDLEEHLLTHPSV
MAB_2247c|M.abscessus_ATCC_199 RSGDRVRALPG----GYLEVTGRIKDVILRGGESIAALDLESHLQTHPAV
MLBr_02661|M.leprae_Br4923 RTGDLGSLSVT----GDLRIRGRIKELINRSGEKISPERVEGVLASHHNV
*:** * : : **:*::* * **.::. :* : :* :
Mflv_2691|M.gilvum_PYR-GCK YAAAAVALPDEYLGEKICAAVVFTGPP---LTLADLNAFLDERGASTHAR
Mvan_3850|M.vanbaalenii_PYR-1 YAAAAVALPDEYLGEKICAAVVFRGKP---ITLAELNAFLDERGASKHAR
TH_2907|M.thermoresistible__bu YAAAAVPLPDEFLGEKICAAVVFRGAP---VSLTELNGYLDERGVATHAR
Rv2384|M.tuberculosis_H37Rv FSAAAVGLPDQYLGEKICAAVVFAGAP---ITLAELNGYLDRRGVAAHTR
Mb2405|M.bovis_AF2122/97 FSAAAVGLPDQYLGEKICAAVVFAGAP---ITLAELNGYLDRRGVAAHTR
MAV_2008|M.avium_104 FSAAAVPLPDPYLGEKICAAVVFEGPA---LSLAELNGYLDERGVAAHAR
MMAR_3698|M.marinum_M RSAAAVPLPDPDMGELICAAIVFAGPPQTPQTLSGLNDYLEQRGVAAHIR
MUL_3641|M.ulcerans_Agy99 RSAAAVPLPDPDMGELICAAIVFAGPPQTPQTLSGLNDYLEQRGVAAHIR
MSMEG_4516|M.smegmatis_MC2_155 WAAAAVALPDEFLGEKICAVVVFNGPP---VSLAELHAHLEQRGVAAHSR
MAB_2247c|M.abscessus_ATCC_199 YAAAAVGLPDQYLGEIVCAAIVFKGKP---VAAAELNQHLQARGAATHSR
MLBr_02661|M.leprae_Br4923 MEVAVFGDPDKVYGETVTAVIVPREVIAP--TPSELAVFCRDR-LAAFEV
.*.. ** ** : *.:* : : * . * : .
Mflv_2691|M.gilvum_PYR-GCK PDVLVPVPSLPTTAVGKVDKKALVAQLTANS---------
Mvan_3850|M.vanbaalenii_PYR-1 PDVLMPVRSLPTTAVGKVDKKQLGAQLTS-----------
TH_2907|M.thermoresistible__bu PDVLVALPSLPMTAVGKVDKKKVVEQVLSQ----------
Rv2384|M.tuberculosis_H37Rv PDQLVAMPALPTTPIGKIDKRAIVRQLGIATGPVTTQRCH
Mb2405|M.bovis_AF2122/97 PDQLVAMPALPTTPIGKIDKRAIVRQLGIATGPVTTQRCH
MAV_2008|M.avium_104 PDVLAAMPSLPTTPIGKIDKKAIANQVSARNRRP------
MMAR_3698|M.marinum_M IDRLVAMPSLPTTPVGKIDKKAIASQILTTTAT-------
MUL_3641|M.ulcerans_Agy99 IDRLVAMPSLPTTPVGKIDKKAIASQILTTTAT-------
MSMEG_4516|M.smegmatis_MC2_155 PDALVPMPSLPTTAVGKIDKKAIVRQLGG-----------
MAB_2247c|M.abscessus_ATCC_199 VDKLVAVSALPLTAVGKIDKRALLASLTG-----------
MLBr_02661|M.leprae_Br4923 PTRFQEASALPHTAKGSLDRRAVAEQFAHRG---------
: :** *. *.:*:: : ..