For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RDIPAELTKRYVEEGWWTPETLGQMLAEGLAAGPDVVFCVHSEVRPYTGTFGEVETLARRLAAGLRARGV GPGDVIAFQLPNWMEAAAAFWATSFLGAVTVPIVHFYGRKELAHILSTAKPKVFLTTERFGRMSFQPDLC AQVPIVGLVGESSFTDLLAERPMSGTLDTDPAAPALIAFTSGTTRDPKGVIHSHQTLSCETRQLLANYPP NRGRQLTATPVGHFIGMVGAFLIPVLEGAPVDLCDVWDPAKVLRLMESDGLSIGGGPPYFVTSLLDHPDF ADRHLARIRTVGLGGSTVPAAVTRRLADLGIFVFRSYGSTEHPSITGSSPSAPEGKRLYTDGNPRPGVEI RLGPDGEIFSRGPDLCLGYTDDALTAKAFDDEGWYRTGDVGFLDDDGYLTITDRTADLIIRGGENISALE VEEVLLGIPGVIEAIVVAAPDARLGERVAAVLRTRSGAPLPTLDDVRAHFERVGIARQKWPEELHHVPDG QDFPRTASGKVQKYRVRQAVRNPAAVMDIATSAN*
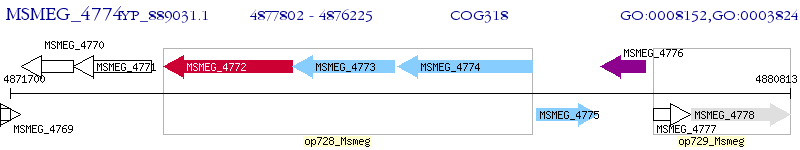
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4774 | - | - | 100% (525) | AMP-dependent synthetase and ligase |
| M. smegmatis MC2 155 | MSMEG_5631 | - | 2e-63 | 36.01% (511) | AMP-dependent synthetase and ligase |
| M. smegmatis MC2 155 | MSMEG_4112 | - | 2e-60 | 31.07% (531) | cyclohexanecarboxylate-CoA ligase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3591 | fadD3 | 3e-49 | 32.99% (488) | acyl-CoA synthetase |
| M. gilvum PYR-GCK | Mflv_2523 | - | 0.0 | 75.05% (521) | AMP-dependent synthetase and ligase |
| M. tuberculosis H37Rv | Rv3561 | fadD3 | 8e-50 | 33.20% (488) | acyl-CoA synthetase |
| M. leprae Br4923 | MLBr_01051 | xclC | 1e-28 | 29.52% (481) | acyl-CoA synthetase |
| M. abscessus ATCC 19977 | MAB_4377c | - | 3e-50 | 31.77% (532) | AMP-binding domain protein |
| M. marinum M | MMAR_0303 | - | 1e-61 | 33.46% (511) | acyl-CoA synthetase |
| M. avium 104 | MAV_2638 | - | 1e-90 | 40.41% (532) | AMP-dependent synthetase and ligase |
| M. thermoresistible (build 8) | TH_2816 | - | 0.0 | 78.68% (516) | PUTATIVE AMP-dependent synthetase and ligase |
| M. ulcerans Agy99 | MUL_4126 | fadD3 | 6e-47 | 32.43% (481) | acyl-CoA synthetase |
| M. vanbaalenii PYR-1 | Mvan_4135 | - | 0.0 | 77.56% (517) | AMP-dependent synthetase and ligase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3591|M.bovis_AF2122/97 ------------------------------MINDLRTVPAALDRLVRQLP
Rv3561|M.tuberculosis_H37Rv ------------------------------MINDLRTVPAALDRLVRQLP
MUL_4126|M.ulcerans_Agy99 ------------------------------MTSDPRTVPTALDRLVRRLP
MAB_4377c|M.abscessus_ATCC_199 --------------------MKAYDAGATLSPIIEETIGENFERIARAHP
MSMEG_4774|M.smegmatis_MC2_155 ---------------MRDIPAELTKRYVEEGWWTPETLGQMLAEGLAAGP
TH_2816|M.thermoresistible__bu ---------------MRSIPAELRRRYEENGWWTQESLGDLLAEGLRAAP
Mflv_2523|M.gilvum_PYR-GCK ---------------MRRIPEELVRRYEQEGYWTPETLGELLARGLDSAP
Mvan_4135|M.vanbaalenii_PYR-1 ---------------MRRIPAQLVERYQREGYWTQETLGDMLARGLAGSP
MAV_2638|M.avium_104 ---------------MPFDHRQLRDKWYSAGWYSDRTCLDAFEAGAVEHK
MMAR_0303|M.marinum_M MGGPKVPQRRLIDGRWVRWDADRAAGAYAHRWWVRRTLADALREAAQSTP
MLBr_01051|M.leprae_Br4923 ------------------------------------MLLASLNPSVVTAT
:
Mb3591|M.bovis_AF2122/97 DHTALIAE---DRRFTSTELRDAVYGAAAALIALGVEPADRVAIWSPNTW
Rv3561|M.tuberculosis_H37Rv DHTALIAE---DRRFTSTELRDAVYGAAAALIALGVEPADRVAIWSPNTW
MUL_4126|M.ulcerans_Agy99 NHDALVTD---ERSFTVSELNDEVYRAAAALIELGVKPAARVGIWSPNTW
MAB_4377c|M.abscessus_ATCC_199 DVGALVDVSG-DRRWTYRELDAEINRVARGLMSLGVAAGDRVGIWAPNCA
MSMEG_4774|M.smegmatis_MC2_155 DVVFCVHSEVRPYTGTFGEVETLARRLAAGLRARGVGPGDVIAFQLPNWM
TH_2816|M.thermoresistible__bu DAGFRVHSDVRPYTGTFADVEERARRLAAGLRARGVGPGDVIAFQLPNWV
Mflv_2523|M.gilvum_PYR-GCK GAGFHVHSAVRPYAGTFAEVERDARRLAAGLRERGVAAGDVVALQLPNWR
Mvan_4135|M.vanbaalenii_PYR-1 EAGFCVHSSIRPYRGTFGEVERDARRLAAGLRERGVGPGDVVAMQLPNWR
MAV_2638|M.avium_104 DVPLTFVIGDSVSSPTVCEIHDRAVALAASLQRLGVRDGDAVAVQLTNRV
MMAR_0303|M.marinum_M QRVALVDADR---QLDCQELHRQAMALAQHLRSR-MPAGSVVSFMLPNWH
MLBr_01051|M.leprae_Br4923 DIADAVCVDG-------VVLSRSGLVGAATSVAERVGGARLVAVLATPTA
. : * : . :.. .
Mb3591|M.bovis_AF2122/97 HWVVACLAIHHAGAAVVPLNTRYTATEATDILDRAGAPVLFAAGLFLGAD
Rv3561|M.tuberculosis_H37Rv HWVVACLAIHHAGAAVVPLNTRYTATEATDILDRAGAPVLFAAGLFLGAD
MUL_4126|M.ulcerans_Agy99 HWVVACLAIHHVGAAMVPLNTRYTADEAADILARTEAPVLFAMGHFLGSD
MAB_4377c|M.abscessus_ATCC_199 EWVLVQYATAKIGAILVNINPAYRTHELAYALNQSGVRTLICAKAFKSSD
MSMEG_4774|M.smegmatis_MC2_155 EAAAAFWATSFLGAVTVPIVHFYGRKELAHILSTAKPKVFLTTERFGRMS
TH_2816|M.thermoresistible__bu EAAMTFWASAFLGAVVVPVVHFYGRKELGHILRTARPKVFITAAEFGRMR
Mflv_2523|M.gilvum_PYR-GCK EAAVTFWASAFLGAVVVPVVHFYGRKELAHILTTAKPRVFVTTAQFGRLQ
Mvan_4135|M.vanbaalenii_PYR-1 EAAVTFWASAFLGAVVVPIVHFYGRRELAHIMATAKPRVFVTAEEFGRMR
MAV_2638|M.avium_104 ECAVAYQAVLLCGAVLVPIVHIYGMAEVAFVLDQSAAKVLIMPERHGSIV
MMAR_0303|M.marinum_M EAAVIYLASTLAAMVANPVLPSLREHELRFILEDAGSRMIFAPADFRGHD
MLBr_01051|M.leprae_Br4923 STVLAITGCLIAGVPVVPVPADIGVVERRHMLTDSGAQAWLGPGPSADSA
. . . : * : : .
Mb3591|M.bovis_AF2122/97 RAAGLDRAALPALRHVVRVPVEADDGTWDEFIATGAGALDAVAARAAAVA
Rv3561|M.tuberculosis_H37Rv RAAGLDRAALPALRHVVRVPVEADDGTWDEFIATGAGALDAVAARAAAVA
MUL_4126|M.ulcerans_Agy99 RVAGLDRKALPALRHIVRIPIEEPDGTWDEFIETGT-DTGAVTARAAAVS
MAB_4377c|M.abscessus_ATCC_199 YVA-MASQVMPDAPGLRDVVFIGTSDWTELVAGAERVTETALRARMSQLS
MSMEG_4774|M.smegmatis_MC2_155 FQPDLC-AQVPIV-GLVG-----------ESSFTDLLAERPMSG-TLDTD
TH_2816|M.thermoresistible__bu YQPDLC-ADVPIV-GVVG-----------QHGFDDLVADEPMTG-TLATD
Mflv_2523|M.gilvum_PYR-GCK FHPDVC-AGVPIV-ALVGDGPDSGADG-GWEPFEDLLAAEPMAG-TVAAD
Mvan_4135|M.vanbaalenii_PYR-1 YQPDLC-GDVAVV-GLVG--AAHPAAG-NEHAFEQLLADEPMAG-TVAAD
MAV_2638|M.avium_104 YSKRLQ-DLSRID-SLQHVVMLDAEPGEGYLAWSQLDTPAARYR-RPAVA
MMAR_0303|M.marinum_M YTSMLRRVAAQLP-SRPEVIEVRGQCPDGVSTFQSLLVGRETAAPLPTAD
MLBr_01051|M.leprae_Br4923 PDG---------------------------LPHIPVQLDARSWNRYPEPS
Mb3591|M.bovis_AF2122/97 PQDVSDILFTSGTTGRSKGVLCAHRQSLSASASWAANGKITSDD--RYLC
Rv3561|M.tuberculosis_H37Rv PQDVSDILFTSGTTGRSKGVLCAHRQSLSASASWAANGKITSDD--RYLC
MUL_4126|M.ulcerans_Agy99 PDDVSDILFTSGTTGRSKGVLCAHRQSLSASASWAANGKITADD--RHLC
MAB_4377c|M.abscessus_ATCC_199 NTDPINIQYTSGTTGYPKGATLSHRNVLNNGFFVAESIQLQAGD--RLCI
MSMEG_4774|M.smegmatis_MC2_155 PAAPALIAFTSGTTRDPKGVIHSHQTLSCETR---QLLANYPPNRGRQLT
TH_2816|M.thermoresistible__bu PAGPALIAFTSGTTRDPKGVIHSHQTLTFETR---QLLANYPPDRGRQLT
Mflv_2523|M.gilvum_PYR-GCK PAGPALIAFTSGTTRDPKGVIHSHQTLSFETR---QLLENYPPDRGRQLT
Mvan_4135|M.vanbaalenii_PYR-1 PAAPTLIAFTSGTTRDPKGVIHSHQTLSCETR---QLLENYPPDRGRQLT
MAV_2638|M.avium_104 ADDVCLLLYTSGTTSAPKGVQHTNNTVLAEQRTMPALLAGKPDD--VSLV
MMAR_0303|M.marinum_M PDAVRMVLYTSGTTGRPKGVLHTHNSIHALICQIRDHWMVEPGD--GFLV
MLBr_01051|M.leprae_Br4923 PDATAMVIYTSGTTGPPKGVLLSRRAIAVDLDALAQAWQWTADD--VLVH
: :***** .**. :.. . :
Mb3591|M.bovis_AF2122/97 INPFFHNFGYKAGILACLQTGATLIPHVT-FDPLHALRAIERHRITVLPG
Rv3561|M.tuberculosis_H37Rv INPFFHNFGYKAGILACLQTGATLIPHVT-FDPLHALRAIERHRITVLPG
MUL_4126|M.ulcerans_Agy99 INPFFHNFGYKAGILACLQTGATLFPHLT-FDPLRTLQAIERHRITVLPG
MAB_4377c|M.abscessus_ATCC_199 PVPFYHCFGMVMGNLGCTTHGATIVMPAPGFDPGRTLEAIERERCVGVYG
MSMEG_4774|M.smegmatis_MC2_155 ATPVGHFIGMVGAFLIPVLEGAPVDLCDV-WDPAKVLRLMESDGLSIGGG
TH_2816|M.thermoresistible__bu ATPVGHFIGMVGAFLIPVLEGAPIDLADV-WDPGRVLRLMEAEDISIGGG
Mflv_2523|M.gilvum_PYR-GCK ATPVGHFIGMLGAFLIPVLEGSPIDLCDV-WDPGRVLQLMETEGLSIGGG
Mvan_4135|M.vanbaalenii_PYR-1 ATPVGHFIGMLGAFLIPVLEGAPIDLCDV-WDPGRVLKLIESDGLSIGGG
MAV_2638|M.avium_104 TFPPGHIAG-VGSMLRPLLSGARTVFMDG-WDPARAVEVVHRFGVTCTAG
MMAR_0303|M.marinum_M ASPIAHIGGSIYAFECPLLLGTTAVLLDR-WDADRAVQLMTSKRCTHMAG
MLBr_01051|M.leprae_Br4923 GLPLFHVHGLVLGLLGSLRIGNRFVHTGK-PTPTAYAQACSEAGGSLYFG
* * * . * . . *
Mb3591|M.bovis_AF2122/97 PPTIYQSLLDHPARKDFDLSSLRFAVTGAATVPVVLVERMQSELDIDIVL
Rv3561|M.tuberculosis_H37Rv PPTIYQSLLDHPARKDFDLSSLRFAVTGAATVPVVLVERMQSELDIDIVL
MUL_4126|M.ulcerans_Agy99 PPTIYQTLLDHPARDDYDLSSLRFAVTGAATVPVVLVERMQSELDIDIVL
MAB_4377c|M.abscessus_ATCC_199 VPTMFIAMLADPGFAHRDLSTLRTGIMAGSVCPVEVMKRCIDEMHMAGVA
MSMEG_4774|M.smegmatis_MC2_155 PPYFVTSLLDHPDFADRHLARIRTVGLGGSTVPAAVTRRLADLG--IFVF
TH_2816|M.thermoresistible__bu PPYFVTSLLDHPDCTPEHLARFKTVGLGGSTVPTTVTRRLAELG--MFVF
Mflv_2523|M.gilvum_PYR-GCK PPYFVTSLLDHPQCRPEHVRRFKTVGLGGSTVPAAVTRRLSDLG--MFVF
Mvan_4135|M.vanbaalenii_PYR-1 PPYFVTSLLDHPDFRPEHLRRFKTVGLGGSTVPAAVTQRLTDLG--LFVF
MAV_2638|M.avium_104 TPFHLAGILDLGDTG-DKLASLREFLVGAAPVAEEIGRRAAQAG--ISTF
MMAR_0303|M.marinum_M ATPFLEQLLAAAQRAGTRLPDLKFFVCGGASVSPSLIRCATDYFDKAVVT
MLBr_01051|M.leprae_Br4923 VPTVWSRLVAD-EAVARALRPARLLVSGSASLPVPVFDRLAHLTG-HRPI
. :: : : ..: . :
Mb3591|M.bovis_AF2122/97 TAYGLTEANGMGTMCRPEDDAVTVATTCGRPFADFELRIADD--------
Rv3561|M.tuberculosis_H37Rv TAYGLTEANGMGTMCRPEDDAVTVATTCGRPFADFELRIADD--------
MUL_4126|M.ulcerans_Agy99 TAYGLTEANGMGTMCRADDDALTVATTCGRPFADFELRIGPDT-------
MAB_4377c|M.abscessus_ATCC_199 IAYGMTETSPVSCQTLFDDDLDRRTATVGRAHPHIEIKIVDPNSGETVQR
MSMEG_4774|M.smegmatis_MC2_155 RSYGSTEHPSITGSS-PSAPEGKRLYTDGNPRPGVEIRLGPD--------
TH_2816|M.thermoresistible__bu RSYGSTEHPSITGSG-PAAPEEKRLYTDGNPRPGVEIRIAED--------
Mflv_2523|M.gilvum_PYR-GCK RSYGSTEHPSITGSR-PSAPEDKRLFTDGDARPGVEIKLGPD--------
Mvan_4135|M.vanbaalenii_PYR-1 RSYGSTEHPSITGSR-PDAPEVKRLFTDGNVRPGVEIRLGED--------
MAV_2638|M.avium_104 RSYGATEHPTVTGEH-EGQPQWARLGTDGKPLPGSEIRIVGSD-GTDCPI
MMAR_0303|M.marinum_M RVYGSTEVPVTTIGA-PDSS-AHAAATDGRVG-AADVKLVAG--------
MLBr_01051|M.leprae_Br4923 ERYGSTESLITLSTL---ADGERRAGWVGLPLAGVQTRLVDESGGPVPYD
** ** * : ::
Mb3591|M.bovis_AF2122/97 ----GEVLLRGPNVMVGYLDDTEATAAAIDADGWLHTGDIGAVDQAGNLR
Rv3561|M.tuberculosis_H37Rv ----GEVLLRGPNVMVGYLDDTEATAAAIDADGWLHTGDIGAVDQAGNLR
MUL_4126|M.ulcerans_Agy99 ----GEVLLRGPNVMLGYLDDPRATAAAIDADGWLHTGDIGVLDEAGNLR
MAB_4377c|M.abscessus_ATCC_199 -GQSGELCTRGYSVMLGYWNDEAHTREVLDTDGWMHTGDLAVMRDDGYCT
MSMEG_4774|M.smegmatis_MC2_155 ----GEIFSRGPDLCLGYTDD-ALTAKAFDDEGWYRTGDVGFLDDDGYLT
TH_2816|M.thermoresistible__bu ----GEILSRGPDLCLGYTDD-ALTARAFDDDGWYHTGDIGVLDDDGYLT
Mflv_2523|M.gilvum_PYR-GCK ----GEIFSRGPDLCLGYVDD-DLTAQAFDEDGWYHTGDIGVLDEDGYLT
Mvan_4135|M.vanbaalenii_PYR-1 ----GEIFSRGPDLCLGYTDD-ELTARAFDDDGWYRTGDVGVLDDDGYLT
MAV_2638|M.avium_104 -GIGGEVVVRGPDQFIGYQDS-ALNTEAFTDDGWFRTGDLGHLDAHGRLT
MMAR_0303|M.marinum_M -----EIRVRGPQMLVGYRHP-EDEAGSFDAQGYFRTGDLGHWVDDEYLV
MLBr_01051|M.leprae_Br4923 GETVGRLQVRSPTMFGGYLNRPEATAEAFDEDGWYRTGDVAVVDSGGMHR
.: *. ** . : :*: :***:.
Mb3591|M.bovis_AF2122/97 INDR-LKDMYICGGFNVYPAEVEQVLARMDGVADAAVIGVPDQRLGEVGR
Rv3561|M.tuberculosis_H37Rv ITDR-LKDMYICGGFNVYPAEVEQVLARMDGVADAAVIGVPDQRLGEVGR
MUL_4126|M.ulcerans_Agy99 ITDR-LKDMYICGGFNVYPAEVEQVLARLDGVADVAVIGIPDHRLGEVGR
MAB_4377c|M.abscessus_ATCC_199 IIGR-LKDMVIRGGENIYPREIEEFLLTHPDIEDVHVVGVPDEKYGEELC
MSMEG_4774|M.smegmatis_MC2_155 ITDR-TADLIIRGGENISALEVEEVLLGIPGVIEAIVVAAPDARLGERVA
TH_2816|M.thermoresistible__bu ITDR-KADLIIRGGENISALEVEEVLLGMPAVAEAVVVAAPDPRLGERTA
Mflv_2523|M.gilvum_PYR-GCK ITDR-KADVIIRGGENISALEVEEILLGMPAVAEAVVVAAPDERLGEHTA
Mvan_4135|M.vanbaalenii_PYR-1 ITDR-KADVIIRGGENISALEVEEVLLAMPAVAEAVVVAAPDARLGEHTA
MAV_2638|M.avium_104 ITDR-IKDVIIRGGETISSGQVEEVLNAHPAVADGVAVAAPDSRYGDVVA
MMAR_0303|M.marinum_M VTGR-AKDIIIRNGENISPKEIEDILITHPGIADVAIVGLPDDRTGERAC
MLBr_01051|M.leprae_Br4923 IVGRESVDLIKLGGYRIGAGEIEMALLGHPDVREVAVVGLPDEDLGQRIV
: .* *: .* : . ::* * : : :. ** *:
Mb3591|M.bovis_AF2122/97 AFVVARPG-TGLDEASVIAYTREH-LANFKTPRSVRFVD---VLPRNAAG
Rv3561|M.tuberculosis_H37Rv AFVVARPG-TGLDEASVIAYTREH-LANFKTPRSVRFVD---VLPRNAAG
MUL_4126|M.ulcerans_Agy99 AFIVARPG-FNLDEKSVIDYTREH-LANFKAPRSVRFVD---TLPRNAGG
MAB_4377c|M.abscessus_ATCC_199 AWVRMRPDRVVIDAVAIRAFASGR-LAHYKIPRYVHVVE---SFPMTVTG
MSMEG_4774|M.smegmatis_MC2_155 AVLRTRSGAPLPTLDDVRAHFERVGIARQKWPEELHHVPDGQDFPRTASG
TH_2816|M.thermoresistible__bu AVVRVKPGHAMPTLDEVRDHFERTGVARQKWPEELHEVS---DYPRTASG
Mflv_2523|M.gilvum_PYR-GCK AVLRIRDGHVMPSLDDVRAHFRAAGAATQKWPEELHRVPEGQDFPRTASG
Mvan_4135|M.vanbaalenii_PYR-1 AVLRIREGQSMPALGDVREHFKQAGVATQKWPEELHRVPEGQDFPRTASG
MAV_2638|M.avium_104 AVVVLKPGYALN-LDDLRAHFAASGLARQKTPERLAIVD---ELPRTSLG
MMAR_0303|M.marinum_M AVIVAAAD-PPPGLPGVRAFLEGAGVARFKIPEQVVIWD---HLPKNHAG
MLBr_01051|M.leprae_Br4923 AFVVGAEA---LDADELINYVAQQ-LSIHKRPREVRFVD---ALPRNAMG
* : : . : * *. : * . *
Mb3591|M.bovis_AF2122/97 KVSKPQLRELG---------------
Rv3561|M.tuberculosis_H37Rv KVSKPQLRELG---------------
MUL_4126|M.ulcerans_Agy99 KVVKPQLRELA---------------
MAB_4377c|M.abscessus_ATCC_199 KVRKIEMRQQTIQIFGLPEPGRTDEQ
MSMEG_4774|M.smegmatis_MC2_155 KVQKYRVRQAVRNPAAVMDIATSAN-
TH_2816|M.thermoresistible__bu KVQKYRVRQSIAEIAAAQQ-------
Mflv_2523|M.gilvum_PYR-GCK KVQKYVIRQQVAAGMHGVVAASQK--
Mvan_4135|M.vanbaalenii_PYR-1 KVQKFVIRQQLAAGELSAIATGQE--
MAV_2638|M.avium_104 KVRKAQLRSDHFGAARR---------
MMAR_0303|M.marinum_M KIVKHRIKAALMKAEQ----------
MLBr_01051|M.leprae_Br4923 KVLKKQLLFEG---------------
*: * :